Figure 1.
SARS-CoV-2 Spike (S) Protein Ectodomain Platform for Characterizing the Structures, Antigenicity, and Protease Susceptibility of the S Protein and D614G Mutant
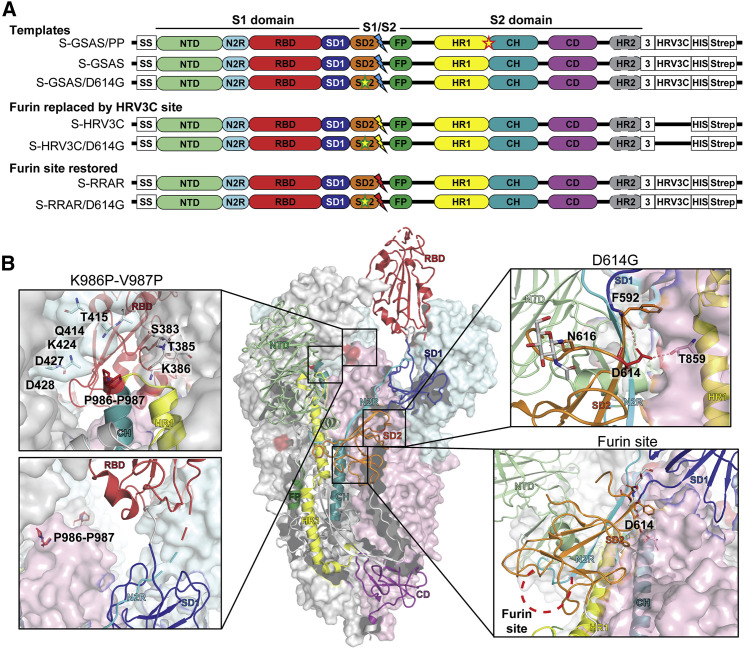
(A) Domain architecture of the SARS-CoV-2 S protomer. The S1 subunit contains a signal sequence (SS), the NTD (N-terminal domain, pale green), N2R (NTD-to-RBD linker, cyan), RBD (receptor-binding domain, red), and SD1 and SD2 (subdomains 1 and 2, dark blue and orange) subdomains. The S2 subunit contains the FP (fusion peptide, dark green), HR1 (heptad repeat 1, yellow), CH (central helix, teal), CD (connector domain, purple), and HR2 (heptad repeat 2, gray) subdomains. The transmembrane domain (TM) and cytoplasmic tail (CT) have been truncated and replaced by a foldon trimerization sequence (3), an HRV3C cleavage site (HRV3C), a his-tag (His), and a strep-tag (Strep). The D614G mutation is in the SD2 domain (yellow star, green contour). The S1/S2 furin cleavage site (RRAR; red lightning) has been mutated to GSAS (blue lightning) or to an HRV3C protease cleavage site (yellow lightning). The K986P-V987P mutations between the HR1 and CH domains are indicated by a yellow star (red contour) on the S-GSAS/PP template.
(B) Representation of the trimeric SARS-CoV-2 S ectodomain with one RBD-up in a prefusion conformation (PDB: 6VSB). The S1 domain on an RBD-down protomer is shown as pale green molecular surface, while the S2 domain is shown in pale red. The subdomains on an RBD-up protomer are colored according to (A) on a ribbon diagram. Each inset corresponds to the S regions understudy and is highlighted in red on the trimeric structure (K986P-V987P, D614G, and the furin protease cleavage site).