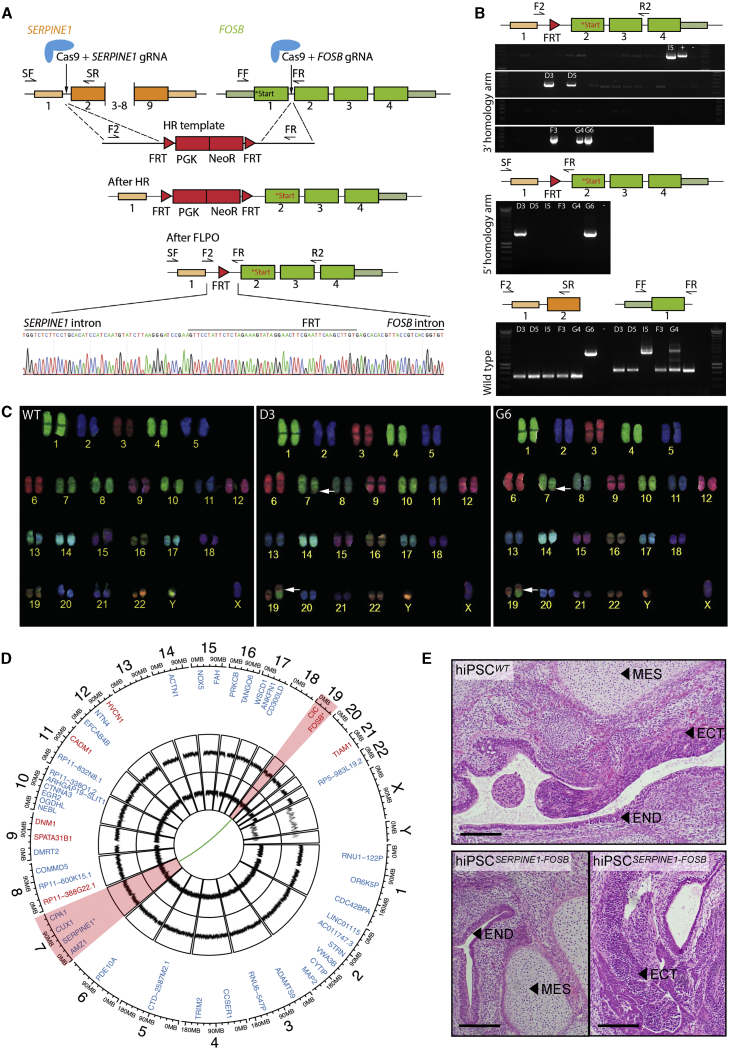
Figure 1.
Generation and Characterization of hiPSCs Carrying the SERPINE1-FOSB Translocation
(A) Schematic overview of the targeting strategy for generation of a SERPINE1-FOSB gene fusion. Filled boxes are exons, lines introns. FOSB start codons are labeled in the figure; black text represents the original start codon, while the new start codon after the fusion is shown in red. Two double-stranded breaks were introduced in the genome guided by 2 gRNAs in SERPINE1 intron 1 and FOSB intron 1. A repair template used for homologous recombination (HR template) with neomycin resistance cassette flanked by Flp-recombinase sequences (FRTs), as well as targeted genomic locus before (After HR) and after FLP-mediated neomycin removal (after FLPO). The bottom panel shows Sanger sequencing of PCR products from the clone with translocation validating HDR recombination of SERPINE1 and FOSB, with the remaining FRT sequence left from the repair template (D3 clone).
(B) Representative results of PCR screen on single-cell-derived hiPSC clones using primers (F2, R2 and SF, SR; F2, SR and FF and FR) shown in the panel above the PCR screen results. Two targeted clones (D3 and G6) were identified of 73 screened clones. PCR shows that clone G6 has a large insert in the SERPINE1 wild-type allele.
(C) COBRA-FISH on colony metaphase cells of WT, D3, and G6 hiPSC clones shows a balanced translocation t(7;19)(q22;q13); furthermore, no additional chromosomal abnormalities were evident in any of the screened cells.
(D) Whole-genome sequencing was performed, and the results are summarized in a Circos plot. The first layer shows all genes that are potential off-target sites for the gRNA for FOSB (red) and SERPINE1 (blue). No mutations were found in the off-target sites and the surrounding 100 bases. The second and third layers show copy number analysis (CNA) for clones D3 and G6, respectively, compared to the isogenic control. No copy number variations (CNVs) are detected. The green connection line shows the detected SERPINE1-FOSB fusion, as detected in both clones D3 and G6. Chromosomes 7 and 19, involved in the translocation, are highlighted in red.
(E) Teratoma formation in mice. The top panel shows teratomas formed from the hiPSCWT, the bottom panel from the hiPSCSERPINE1-FOSB (D3); 2 sections of each are shown. Cellular derivatives of the 3 germ lineages are indicated: mesoderm (MES), ectoderm (ECT), and endoderm (END). Scale bar indicates 200 μm.