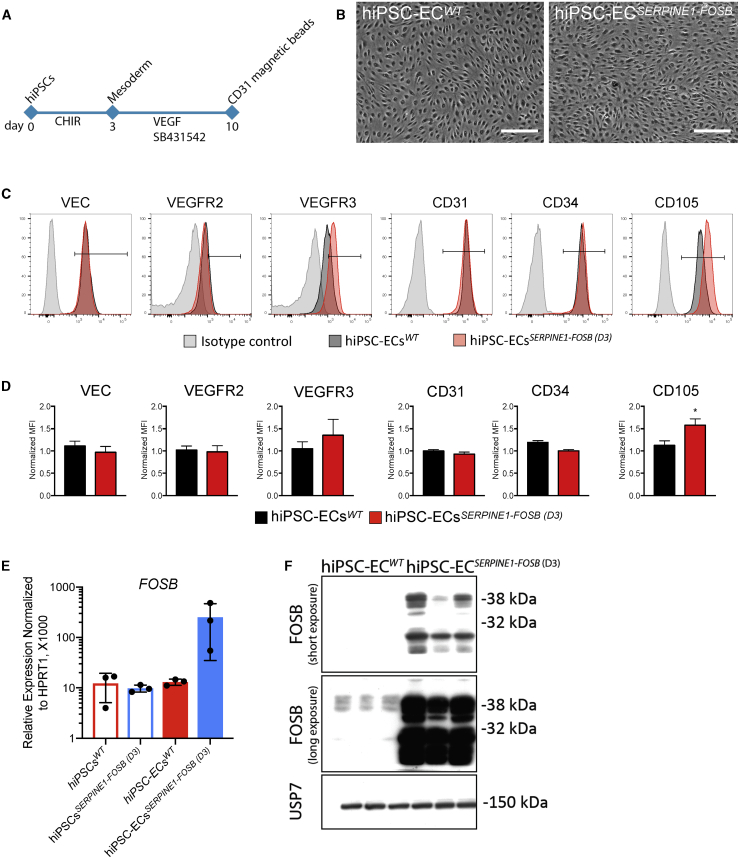
Figure 2.
hiPSC-ECs Carrying the SERPINE1-FOSB Translocation Show Increased FOSB Expression
(A) Schematic overview of the differentiation protocol and purification of ECs from hiPSCs.
(B) Bright-field images showing typical EC morphology of hiPSC-ECs. Scale bar represents 500 μm.
(C) Fluorescence-activated cell sorting (FACS) analysis of EC marker expression on isolated ECs at passage 3 (P3) from hiPSC-ECsWT (black-filled histogram) and hiPSC-ECsSERPINE1-FOSB (D3) (red-filled histogram), and relevant isotype control (gray-filled histogram).
(D) Quantification of normalized relative surface expression levels (MFI) of VEC, VEGFR2, VEGFR3, CD31, CD34, and CD105. n = 3 (biological replicates, 3 independent batches of hiPSC-ECs). Error bars are SDs, ∗p < 0.005.
(E) Real-time qPCR analysis of FOSB expression in hiPSCsWT, hiPSCsSERPINE1-FOSB (D3), hiPSC-ECsWT, and hiPSC-ECsSERPINE1-FOSB (D3) normalized to the housekeeping gene HPRT1 (×1,000). n = 3 (biological replicates, 3 independent batches of hiPSC-ECs). Error bars represent means ± SDs.
(F) Western blot of FOSB expression in hiPSC-ECsWT and hiPSC-ECsSERPINE1-FOSB (D3). Short and long exposure of the gel is shown. USP7 was used as a housekeeping control.