Figure 3.
Transcriptome Analysis of hiPSC-ECs Carrying the SERPINE1-FOSB Translocation
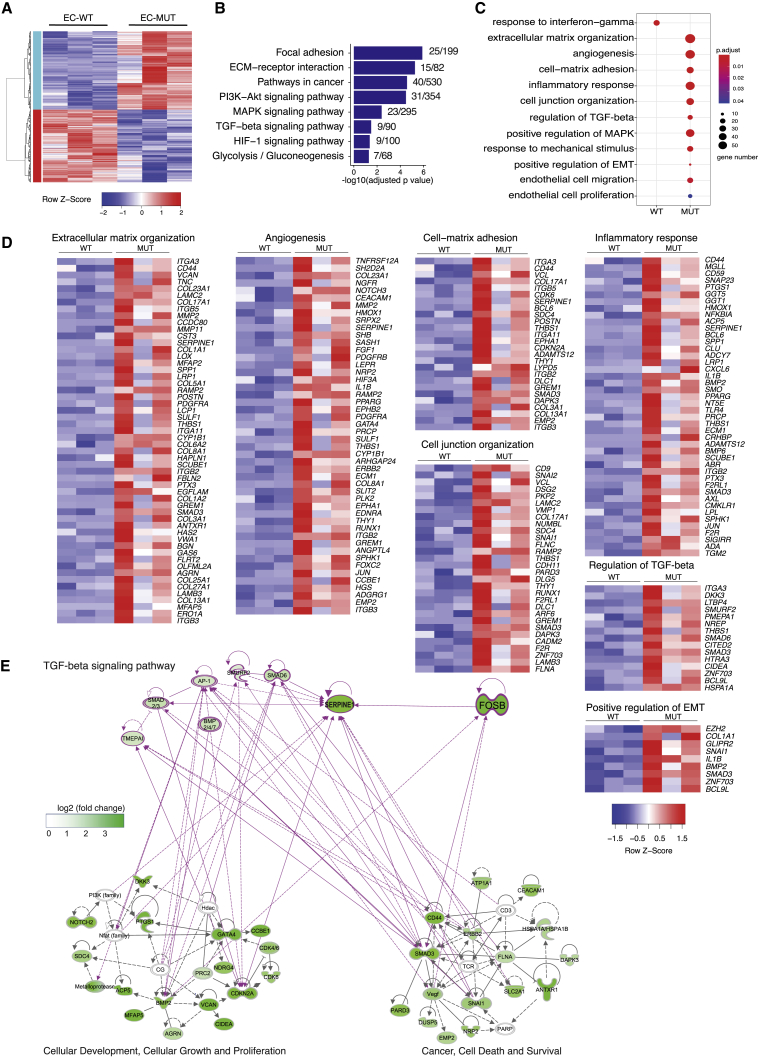
(A) Hierarchical clustering analysis (HCA) of differentially expressed genes (DEGs) between hiPSC-ECsWT (WT) and hiPSC-ECsSERPINE1-FOSB (D3) (MUT) samples (3 independent differentiations and isolation for each clone). A total of 630 and 592 significantly upregulated and downregulated genes in MUT were identified compared to WT ECs (pFDR ≤ 0.05).
(B) Representative KEGG pathways enriched in DEGs upregulated in MUT ECs (−log10(adjusted p value)) and number of enriched genes within total genes of each pathway are shown.
(C) Representative Gene Ontology (GO) enriched in DEGs upregulated in WT or DEGs upregulated in MUT ECs. Size and color indicate gene number and adjusted p value of each GO.
(D) Heatmaps of genes from GOs enriched in hiPSC-ECsSERPINE1-FOSB(D3) upregulated DEGs.
(E) Gene interaction network of genes from GOs shown in (C and D and Figure S4C) constructed using Ingenuity Pathway Analysis (IPA). SERPINE1 and FOSB were added manually. Interactions among FOSB and TGF-β signaling pathway and 2 networks were generated using IPA. Color indicates the log2(fold change) of gene expression in MUT compared to WT.