Figure 1.
Innate Immune Response in COVID-19 Patients at the Time of Admission
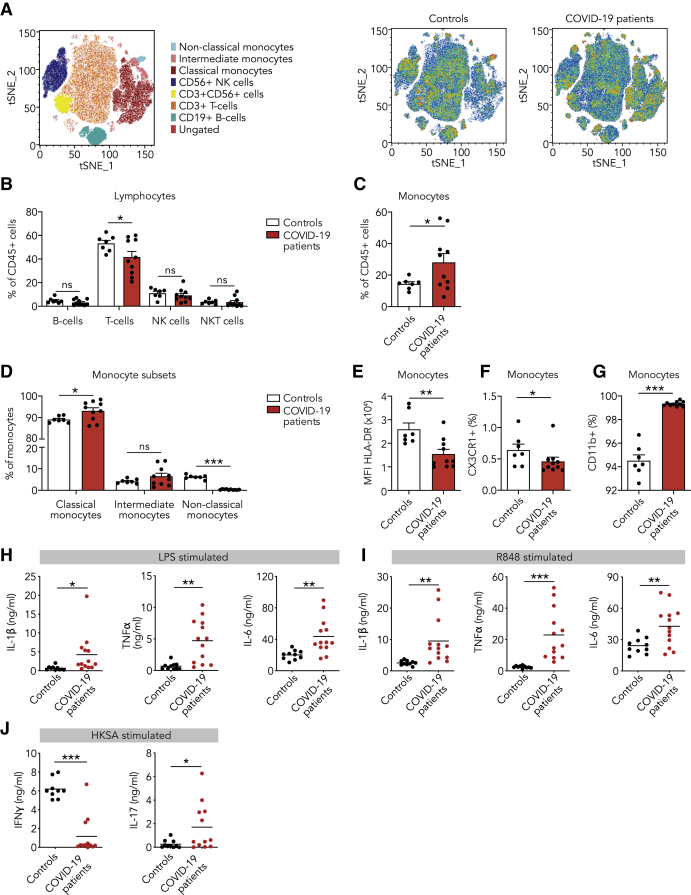
(A–G) PBMCs isolated from COVID-19 patients at admission and from healthy controls were analyzed using flow cytometry (n = 10 for COVID-19 patients, n = 7 for healthy controls).
(A) t-distributed stochastic neighbor embedding (tSNE) plots showing unsupervised clustering on the expression of 10 markers (CD45, CD14, CD16, CD3, CD19, CD56, HLA-DR, CD11b, CCR2, and CX3CR1) in controls and COVID-19 patients.
(B) Quantification of lymphocytes using gating strategy shown in Figure S1C indicated decreased amounts of T cells in COVID-19 patients.
(C and D) Quantification of monocytes showed overall higher counts in COVID-19 patients that was due to higher number of classical monocytes (CD142+,CD16−), whereas non-classical monocytes (CD14+, CD162+) were reduced in COVID-19 patients.
(E–G) Analysis of marker expression on monocytes revealed reduced expression of HLA-DR (E), reduced number of CX3CR1-expressing monocytes (F), and increased number of CD11b-expressing monocytes (G) in COVID-19 patients.
(H and I) Isolated PBMCs were stimulated with LPS (H) or R848 (I) for 24 h, after which the production of IL-1β, IL-6, and TNF-α was quantified in the supernatant using ELISA. COVID-19 patient PBMCs show increased cytokine production upon stimulation with either stimulus (n = 13 for COVID-19 patients, n = 10 for healthy controls).
(J) Isolated PBMCs were stimulated with heat-killed Staphylococcus aureus (HKSA) for 7 days, after which the production of IFNγ and IL-17 was quantified using ELISA. IFNγ response was reduced, whereas IL-17 production was elevated in COVID-19 patients. (n = 12 for COVID-19 patients, n = 10 for healthy controls)
Data are presented as mean ± SEM.∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 for two-sided Student’s t test (for normally distributed data) or Kruskal-Wallis test.