Figure 2.
Innate Immune Response in COVID-19 Patients at Presentation and the Relation to Outcome
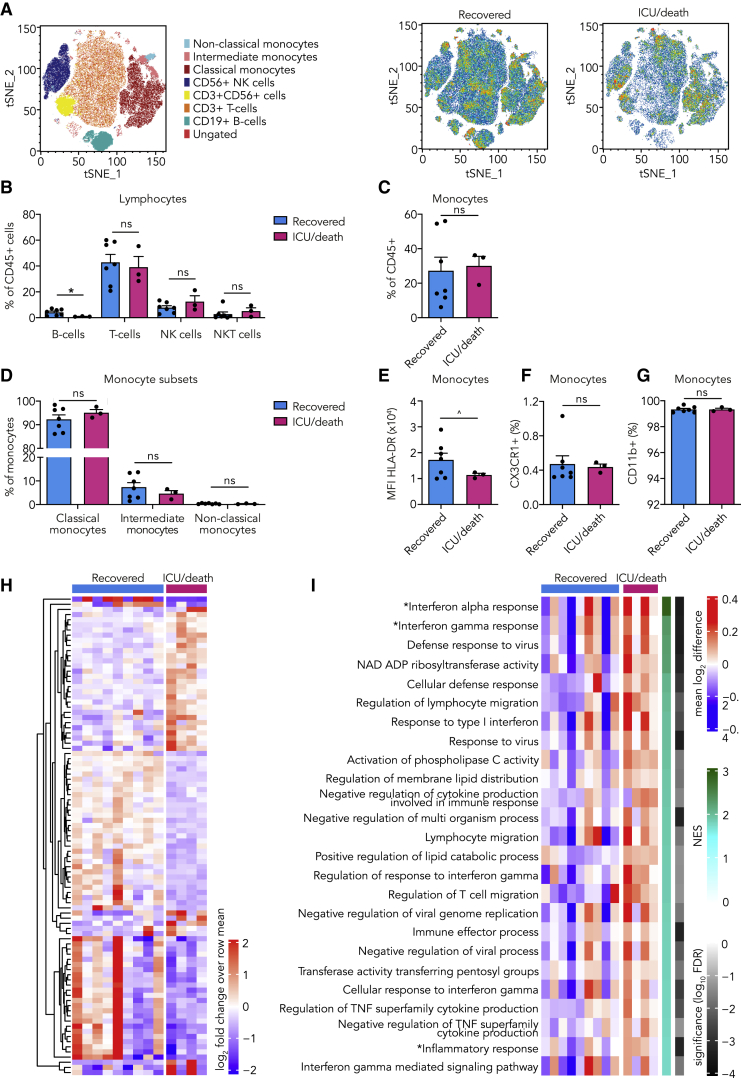
(A–G) PBMCs isolated from COVID-19 patients at admission were analyzed using flow cytometry (n = 7 for COVID-19 patients who recovered, n = 3 for COVID-19 patients who eventually required ICU admission or died).
(A) tSNE plots showing unsupervised clustering on the expression of 10 markers (CD45, CD14, CD16, CD3, CD19, CD56, HLA-DR, CD11b, CCR2, and CX3CR1) in COVID-19 patients who recovered versus those who eventually required ICU admission or died.
(B) Quantification of lymphocytes using gating strategy shown in Figure S1C indicated minor differences in B cells between COVID-19 patients who recovered versus those who eventually required ICU admission or died.
(C and D) Quantification of monocytes showed no difference between COVID-19 patient groups.
(E–G) Expression of HLA-DR (E), CX3CR1 (F), and CD11b (G) on monocytes did not differ between COVID-19 patient groups.
(H and I) Transcriptome analysis was performed on isolated monocytes of COVID-19 patients at admission. (n = 9 for COVID-19 patients who recovered, n = 4 for COVID-19 patients who required ICU admission or died)
(H) Heatmap of differentially expressed genes (p < 0.005) (listed in Table S3) between COVID-19 patients who recovered versus those who eventually required ICU admission or died.
(I) Heatmap of Gene Ontology (GO) pathways and hallmark pathways that are significantly enriched (false discovery rate [FDR], <0.05) in COVID-19 patients who recovered versus those who eventually required ICU admission or died (∗, hallmark pathways; NES, normalized enrichment score).
Data are presented as mean ± SEM; ˆp < 0.06, ∗p < 0.05 for two-sided Student’s t test (for normally distributed data) or Kruskal-Wallis test.