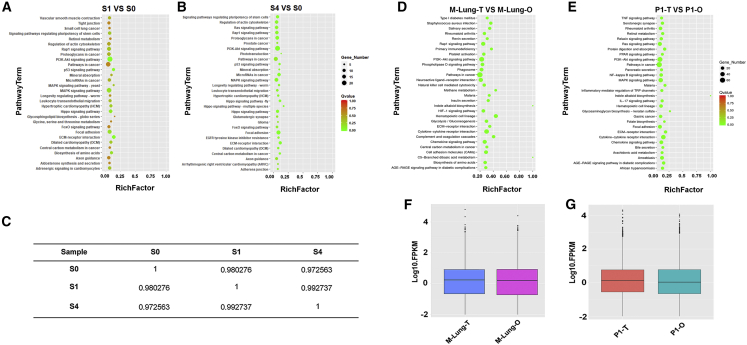
Figure 4.
Cell-Phenotype Consistency of Organoids and Their Parental Tissues/Tumors Profiled by RNA-Seq
(A and B) Scatterplot of the KEGG pathway enrichment analysis of differentially expressed genes in paired comparisons of (A) S0 versus S1 and (B) S0 versus S4 iPSCs. The Rich factor in the x axis is the ratio of differentially expressed gene numbers annotated in a pathway term to all gene numbers annotated in this pathway term. A greater Rich factor indicates a higher degree of pathway enrichment. The color codes the p values.
(C) Pairwise Spearman’s ρ correlation coefficients between S0, S1, and S4 iPSCs. The iPSCs were loaded in Matrigel at 1.0 × 107 cells mL–1, formulated into droplets, incubated in tubing for 10 min, printed, and cultured in vitro before being sequenced.
(D and E) Scatterplot of the KEGG pathway enrichment analysis of differentially expressed genes in paired comparisons of (D) mouse lung tissue (M-Lung-T) versus mouse lung organoid (M-Lung-O) and (E) patient 1 derived organoid (P1-O) versus patient 1 tumor (P1-T, lung).
(F and G) Boxplot of the log FPKM expression values in (F) M-Lung-T and M-Lung-O, and (G) P1-O and P1-T.