Figure 1.
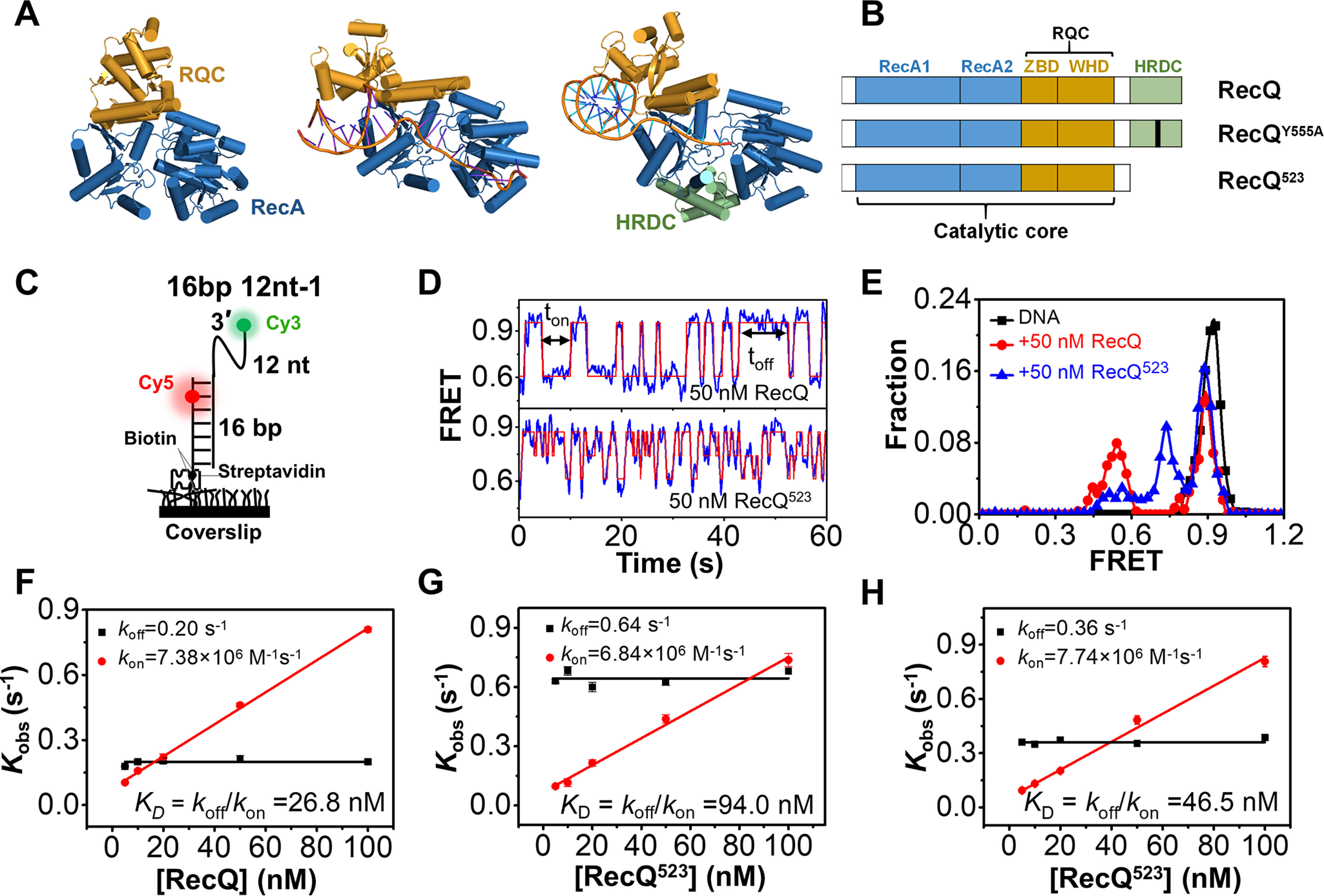
RecQ helicase repetitively associates with and dissociates from the 3′-partial duplex DNA. A, crystal structures of an HRDC-deleted E. coli RecQ (PDB: 1OYW), HRDC-deleted C. sakazakii RecQ in complex with DNA (PDB: 4TMU), and HRDC-containing human BLM in complex with DNA (4O3M). B, domain map of E. coli RecQ constructs used in this study. C, schematic set-up of the smFRET experiment. D, the typical smFRET traces of 16 bp 12 nt-1 in 50 nm RecQ and RecQ523. An automated step-finding algorithm was used to identify the different FRET states (red line). E, FRET histograms of 16 bp 12 nt-1 in 50 nm RecQ or RecQ523 based on the fitted FRET traces. In all the following figures, the FRET histograms were collected from more than 200 traces. F–H, the dissociation rate (koff =1/ton′, red) and the binding rate (kon=1/toff′, black) as a function of protein concentrations. As expected for a binary reaction, the dissociation rate is independent of protein concentration whereas the binding rate has a linear dependence on it. The dissociation constant is thus determined as KD = koff/kon. Error bars denote the standard deviations.
