Figure 7.
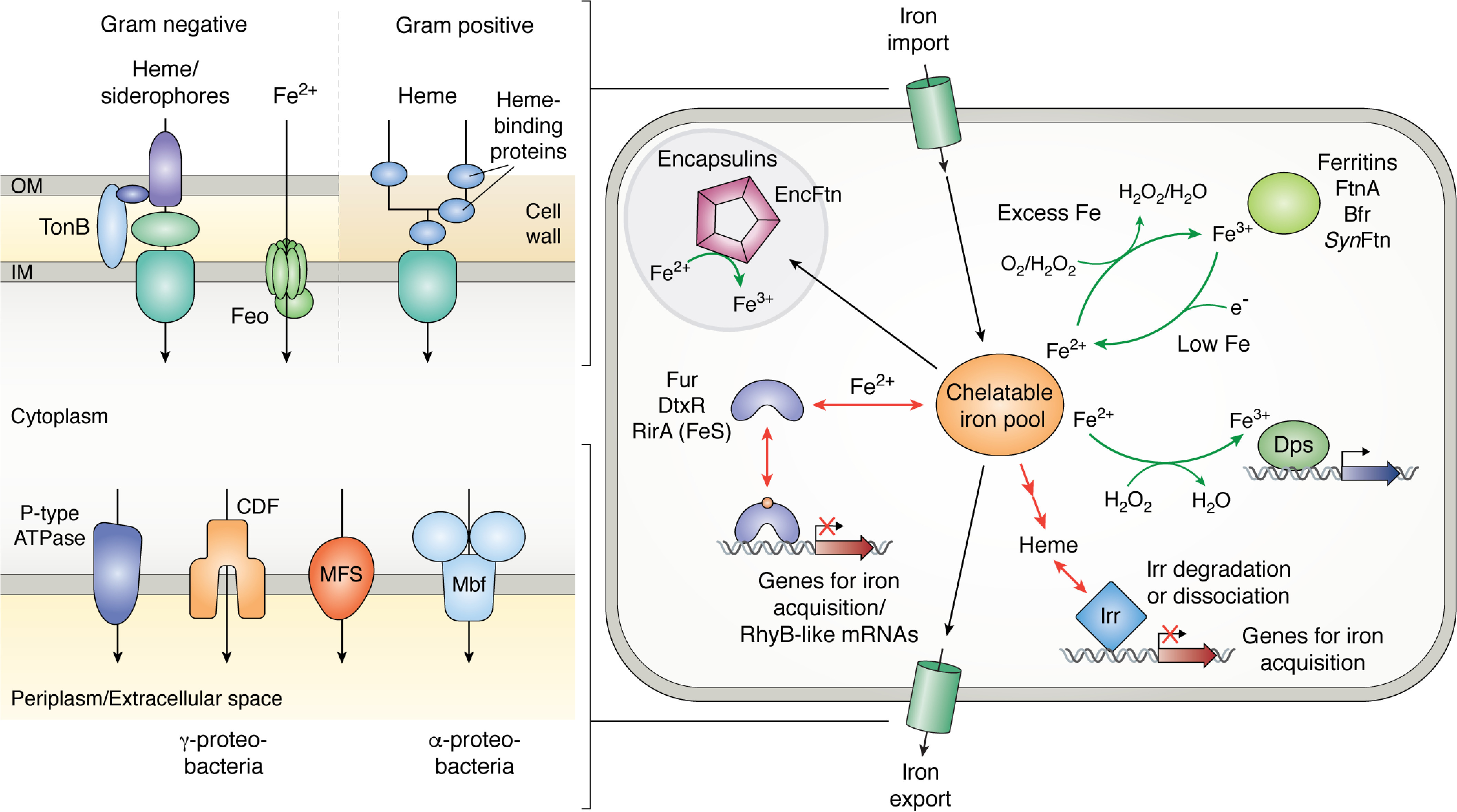
Schematic overview of the major components of iron sensing and detoxification found in bacterial cells. Note that not all of these components are present in a single bacterial cell. Regulatory proteins are shown here as repressors but, in some cases, can also act as activators. Encapsulins are large protein compartments that house EncFtn ferritin-like proteins. The fate of iron stored in encapsulins and in Dps proteins is not clear, although it is likely that at some point, it becomes bioavailable again. Ftn, Bfr, and Dps do not appear to be distributed according to phyla. Fur is the transcriptional regulator in most bacteria but is replaced by DtxR/IdeR in some actinobacteria. In the α-proteobacteria, Fur plays a diminished role in iron homeostasis, with the majority of these functions being performed by Irr. In some rhizobiales, this is achieved in conjunction with a second global regulator, RirA. Import of siderophores and heme across the cytoplasmic membrane (IM) is performed by ABC transporters in all known cases, and Feo is the major importer of Fe2+. In Gram-negative bacteria, heme and siderophores are imported to the periplasm by outer-membrane (OM) porins, whereas a network of heme-binding proteins transports this cofactor across the cell wall of the Gram-positive bacteria. Characterized Fe2+ export systems are rare, but P-type ATPases are the most widely distributed. IceT of S. typhimurium is the only example of the MFS characterized to date, whereas the CDF proteins are limited to γ-proteobacteria and the MbfA proteins to α-proteobacteria. YiiP from E. coli is the only Fe2+ efflux pump for which the structure has been solved (227).
