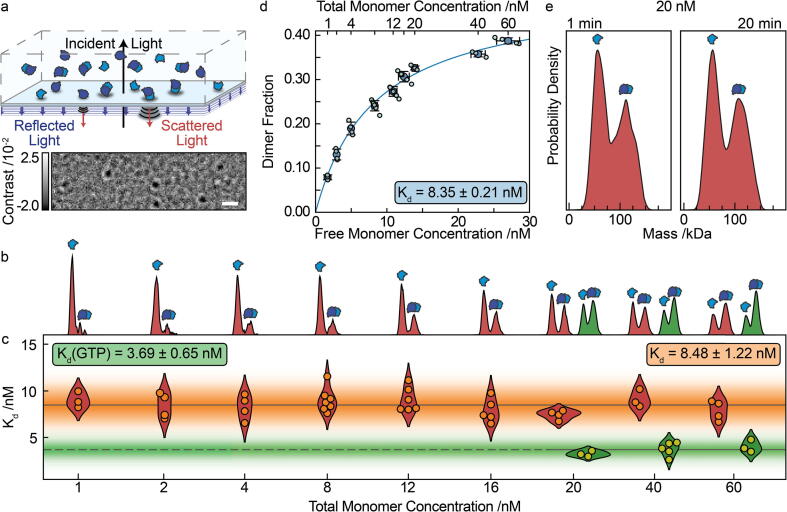
Figure 1.
Quantification of tubulin heterodimersiation with mass photometry. (a) Schematic illustrating the operation of mass photometry. Imaging the interference between scattered and reflected light as a protein non-specifically binds at a glass-water interface results in label-free single molecule images with a contrast proportional to their molecular mass. Scale bar: 1 µm. (b) Mass kernel density estimates with 5 kDa bandwidth for tubulin at total monomer concentrations ranging from 1 to 60 nM without (red) and 20 to 60 nM with (green) GTP incubation. (c) Resulting binding affinities extracted from each distribution in (b). The grey lines indicate the global mean and the shaded areas the standard deviations for each data set. (d) Proportion of dimer present without GTP incubation at equilibrium as a function of free monomer concentration at equilibrium. Light blue markers indicate individual experiments, dark blue markers and error bars depict the mean and standard deviation respectively for each total monomer concentration. A logistic fit through the mean values yields = 8.35 ± 0.21 nM (error on fit). (e) Averaged mass kernel density estimates (N = 4) with 5 kDa bandwidth for tubulin without GTP incubation at a total monomer concentration of 20 nM from separate experiments 30 s after (left) and 20 min after dilution (right).