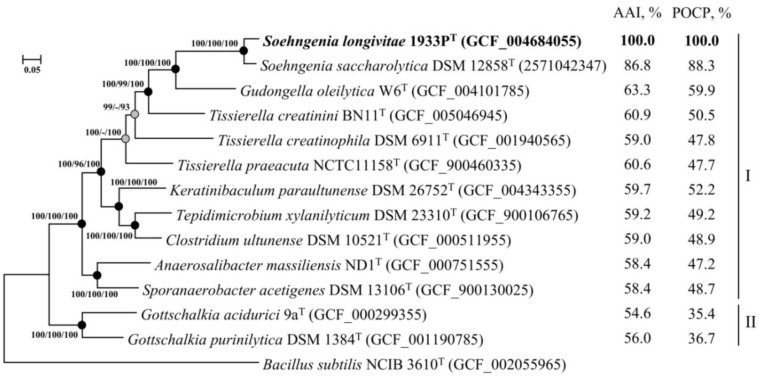
Figure 3.
Maximum-likelihood phylogenetic tree derived from concatenated 120 single copy marker proteins showing the position of strain 1933PT in relation to taxonomically characterized members of the families Tissierellaceae (I) and Gottschalkiacecae (II). Phylogenetic analysis was performed with an LG + F + I + G4 model based on 34,747 amino acid positions. Scale bar, 0.05 amino acid substitutions per site. The tree was rooted using Bacillus subtilis NCIB 3610T as the outgroup. Accession numbers for the genomes are indicated in brackets. The average amino acid identity (AAI) and pairwise percentage of conserved proteins (POCP) values refer to strain 1933PT compared to the rest of the species.