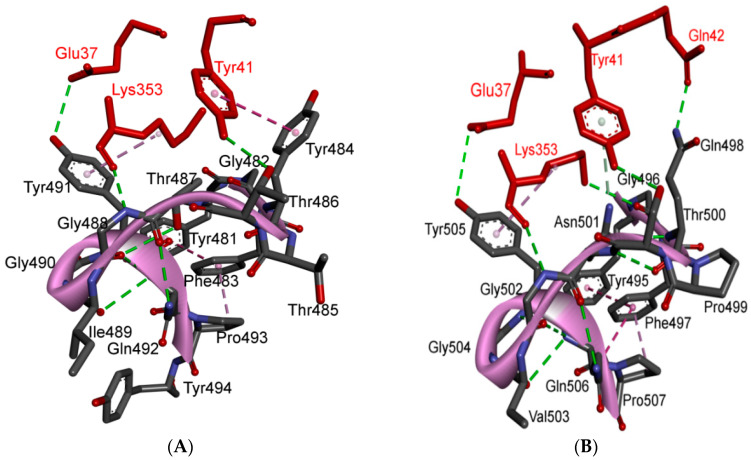
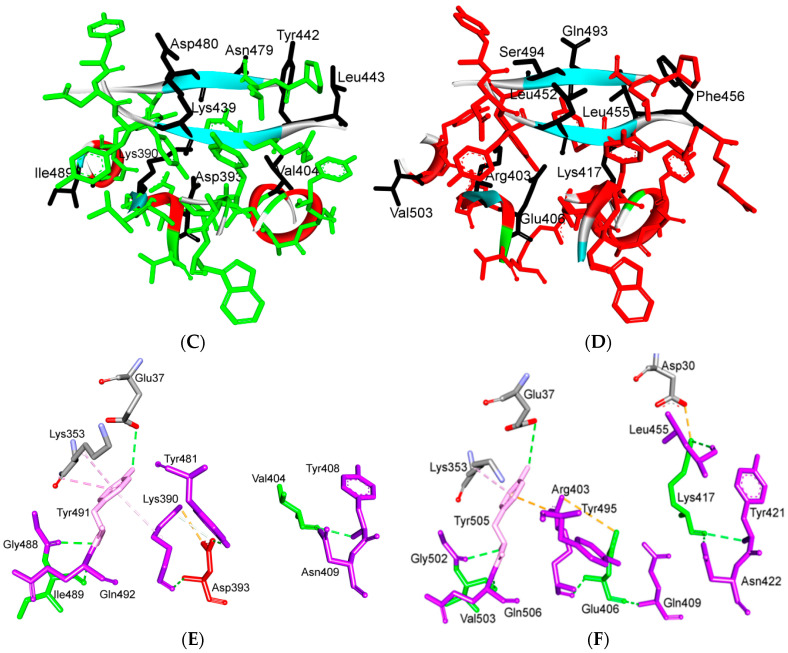
Figure 5.
Structure of TR2 and ridge regions in both SARS-CoV and SARS-CoV-2, and their interactions to ACE2. (A,B) show the intra-molecular and inter-molecular contacts of TR2 region in SARS-CoV and SARS-CoV-2, respectively. The number of the interactions in TR2 region is noticeably higher in SARS-CoV-2 compared to that of SARS-CoV. The presence of Asn501 in SARS-CoV-2 (instead of Thr487) makes a substantial difference since it is involved in stacking with Tyr41 on ACE2. This stacking liberates Gln498 (instead of Tyr484 in SARS-CoV) to make a strong hydrogen bond with Gln42 of ACE2. (C,D) show the environment of the ridge and upper core regions of SARS-CoV and SARS-CoV-2, respectively. The residues that are labelled in black show the amino acids in this region that differ between SARS-CoV and SARS-CoV-2. (E,F) show the main amino acids in the upper core region. The green and red residues indicate those in α-helices and 3/10 helices in this region, respectively. These amino acids are in contact with other amino acids in the RBM (pink) and RBD (violet). Lys417 is the main player in this region, since it is involved in electrostatic interactions with Asp30 from ACE2. 3D models were created using Discovery Studio (version 2.5.5, Biovia, San Diego, CA, USA).