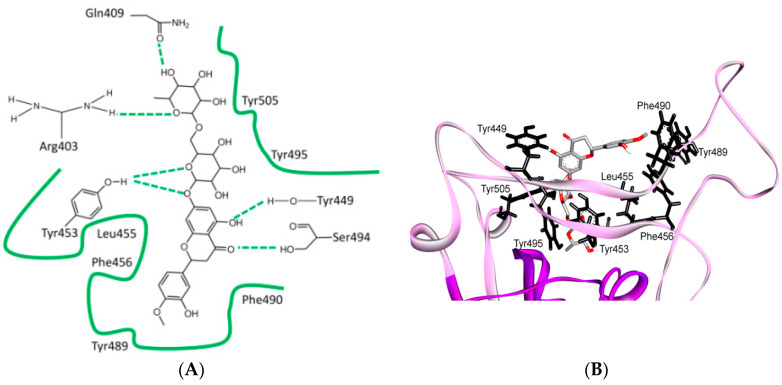
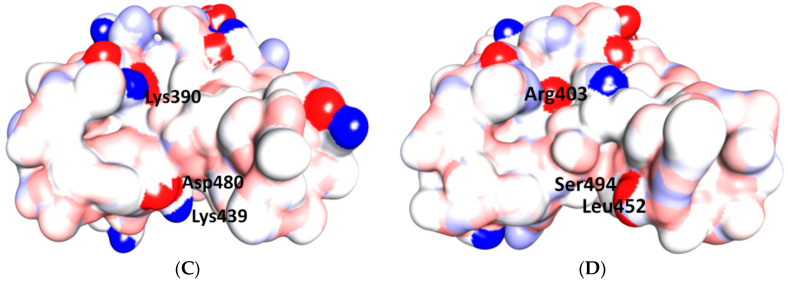
Figure 6.
Differences of shallow pit in the ridge region of RBD for SARS-CoV and SARS-CoV-2, and its binding with hesperidin in SARS-CoV-2. (A) Two-dimensional structure of hesperidin and its surrounding interactions with the SARS-CoV-2 RBD. (B) Hesperidin has been predicted to lie on the middle shallow pit of the surface of the RBD of the S protein, where the dihydroflavone part of the compound lies parallel with the β6 sheet of the RBD. The sugar moiety is inserted into the shallow pit in the direction away from ACE2, where a few hydrophobic amino acids, including Tyr449, Try453, Leu455, Phe456, Phe490, Try489, Try495 and Tyr505 (corresponding to Tyr436, Tyr440, Tyr442, Leu443, Trp476, Tyr475, Tyr481 and Tyr491 in SARS-CoV) form a relatively hydrophobic shallow pocket to contain the compound. Hydrogen-bonding interactions have been predicted between Tyr453, Ser494, Gln409, Tyr449, Arg403 (corresponding to Tyr440, Asp480, Gln396, Tyr449 and Lys390 from SARS-CoV) and the compound. Tyr453 and Tyr449 are among the critical amino acids. (C,D) in the space-filling models of the S protein in SARS-CoV (left) and SARS-CoV-2 (right), the shallow pit is apparent (Tyr453 and Tyr449 are among the critical amino acids). The hydrophobic cleft is apparently similar in both viruses; the differences are mainly located at the gates of this cleft, where Asp480 and Lys439 at one entrance in SARS-CoV are replaced by Ser494 and Leu452 in SARS-CoV-2, while Lys390 at the other entrance in SARS-CoV is replaced by Arg403 in SARS-CoV-2. These changes may affect the binding affinities of proposed drugs with SARS-CoV and SARS-CoV-2. It has been reported that the electrostatic potential observed for SARS-CoV-2 is more positive than that for SARS-CoV, which is consequently considered to result in a greater electrostatic interaction between ACE2 and SARS-CoV-2. This could be explained by the replacement of the negatively charged amino acid Asp by a neutral Ser (Asp480 to Ser494) [59]. Three-dimensional models were created using Discovery Studio (version 2.5.5, Biovia, San Diego, CA, USA).