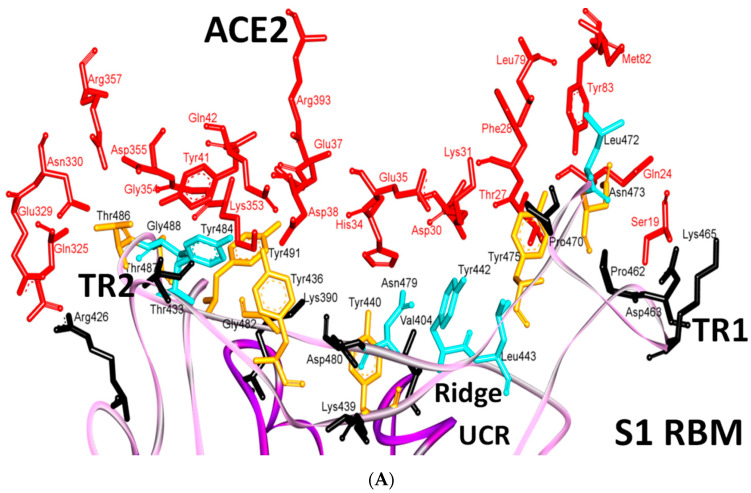
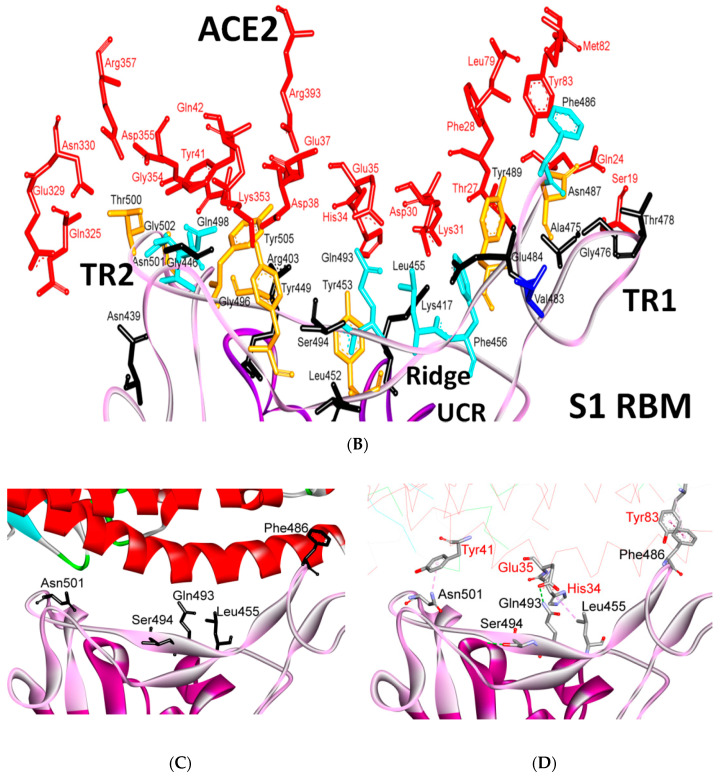
Figure 11.
Binding interface and interactions between RBM of SARS-CoV and SARS-CoV-2 with ACE2. (A,B) interacting residues between the RBM and ACE2 for SARS-CoV and SARS-CoV-2, respectively. Red, orange, cyan and black colors indicate the interacting amino acids from ACE2, identical residues between SARS-CoV and SARS-CoV-2, different residues with partially conserved contacts, and different residues with different contacts, respectively. Ridge: region between TR1 and TR2. UCR: Upper core region. A Leu to Phe486 mutation (in addition to Pro to Ala475, Pro to Glu484, Lys to Thr478, Asp to Gly476 mutations and a Val483 insertion) in TR1 increases its hydrophobicity and the order of its structure, while mutations in the ridge region of Val to Lys417 (which makes electrostatic interactions), Asn to Gln493 (more hydrogen bonding), Asp to Ser494 and Lys to Leu452 (which increase the hydrophobicity of the shallow pit) give SARS-CoV-2 an advantage to bind mainly through these regions. Arg426, Thr433 and Tyr484 in SARS-CoV (instead of Asn439, Gly446 and Gln498 in SARS-CoV-2) produce more electrostatic, hydrogen bonding and stacking interactions, respectively, which give SARS-CoVan advantage to bind mainly through TR2. The use of different regions for binding may explain the differences in affinity and pathogenesis between SARS-CoV and SARS-CoV-2. (C) shows the five critical binding residues that are shown in Table 2. (D) shows the interacting residues from ACE2 with these critical residues. Three-dimensional models were created using Discovery Studio (version 2.5.5, Biovia, San Diego, CA, USA).