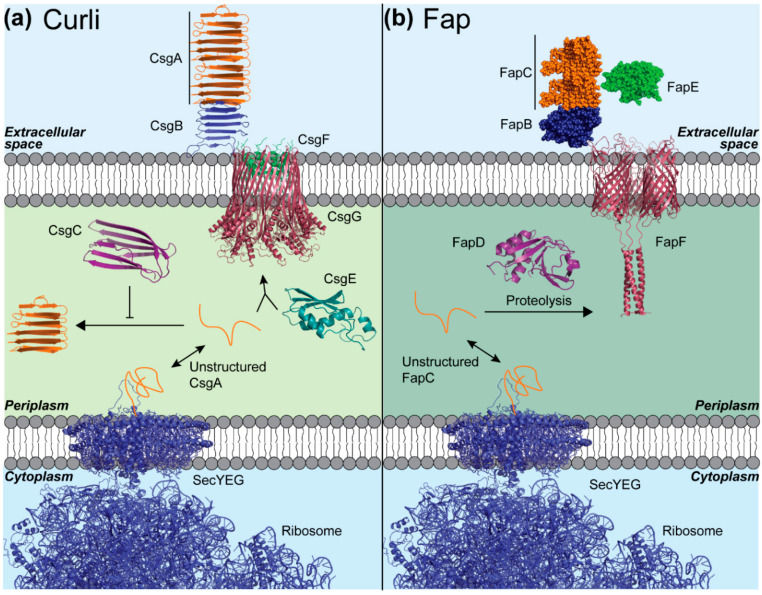
Figure 3.
The bacterial amyloid systems called curli in E. coli and fap in P. aeruginosa are controlled through complex, multi-protein mechanisms. (a) The curli operon contains 7 proteins (6 shown here), each playing a necessary role in maintaining spatiotemporal control over polymerization of the major curli subunit CsgA [72]. CsgA is translated and translocated directly into the periplasm using the SecYEG secretion pore (PDB: 4V6M). CsgC (PDB: 2y2y) is a chaperone-like protein which inhibits CsgA aggregation within the periplasm. CsgE (PDB: 2NA4) is another periplasmic chaperone which fosters CsgA translocation through the nonameric curli assembly pore CsgG (PDB: 6L7A). Lastly, CsgF (PDB: 6L7A) and CsgB (REF [73]) both help to localize curli formation to the CsgG pore and the outer membrane, respectively. (b) In Pseudomonas, the major fap component FapC, is secreted into the periplasm using a SecYEG pore. FapD (modeled after the homologous C39 peptidase domain of ABC transporter PCAT1, PDB: 4RY2) is a peptidase which performs an essential proteolytic modification to one or more of the fap proteins. FapC is passed through the outer membrane using FapF, a trimeric polypeptide transporter (PDB: 5O67). Finally, FapB and FapE are essential minor components of fap amyloids, with FapB potentially playing a nucleator role similar to CsgB. Models shown of FapC, FapB, and FapE are structural predictions produced by the FALCON@home server, since there is no putative structural data in the literature.