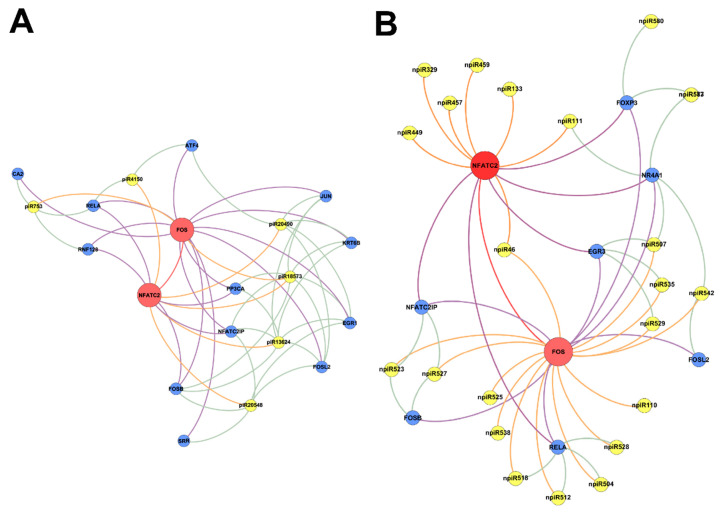
Figure 4.
Networks of differentially expressed piRNAs targeting FOS and NFATC2 operating during PHCM challenge with T. cruzi. Biological interaction networks were created using predicted piRNA target genes (FOS, NFATC2) as primary seed nodes to query pathway and interaction data sources out to one degree of interaction. Primary seed nodes are displayed as red circles. Expansion nodes (blue circles) with a predicted binding target of a differentially expressed known or novel piRNA (yellow node) were added to the network. Connections between nodes (edges) represent interactions between the different biological entities. Networks of (A) known piRNAs and (B) novel piRNAs targeting FOS, NFATC2 and adjacent genes from expansion to one degree of interaction during all time points.