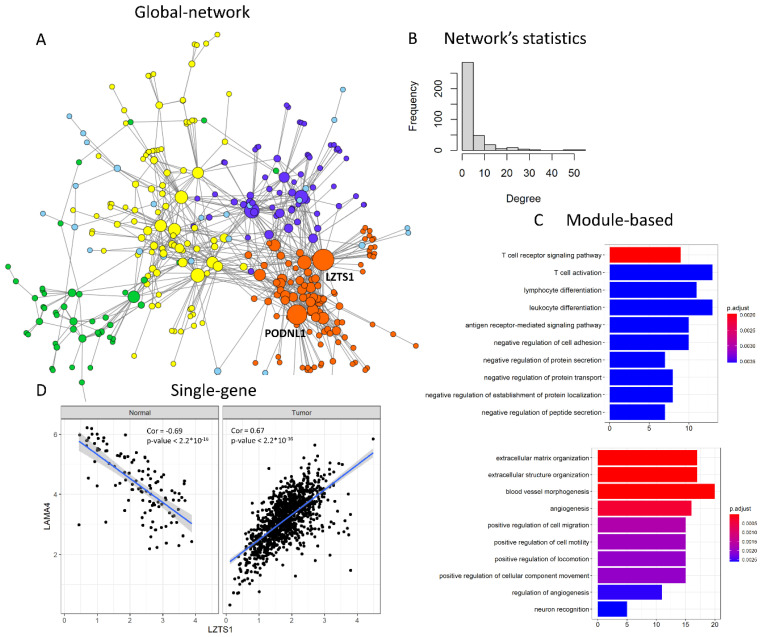
Figure 4.
Example of differential co-expression analysis comparing TCGA normal tissue and breast tumour transcriptional data. (A) In a “global network” approach, the whole differentially co-expressed network is obtained. Here, only the top significantly differentially co-expressed genes are shown for simplicity (adjusted p-value < 10−40). Each node is a gene, colour indicates the cluster and size indicates the number of connections of that gene. The two nodes with the highest degree of differential connections are indicated (LZTS1 and PODNL1); (B) The differentially co-expressed network can be analysed as a whole, studying its topological properties, such as degree distribution; (C) In the global network, clusters (modules) can be identified and studied through functional enrichment, in a “module-based” approach. The top significantly enriched Gene Ontology (GO) categories for the two largest modules are shown in the two boxes, with bars indicating the number of genes belonging to each GO category and colour indicating the adjusted p-value; (D) Node and edge prioritisations allow for selecting specific genes or gene pairs with a particularly strong change in connectivity between the compared conditions: LZTS1 and LAMA4 are significantly negatively correlated in normal breast and switch to a positive correlation in breast tumours.