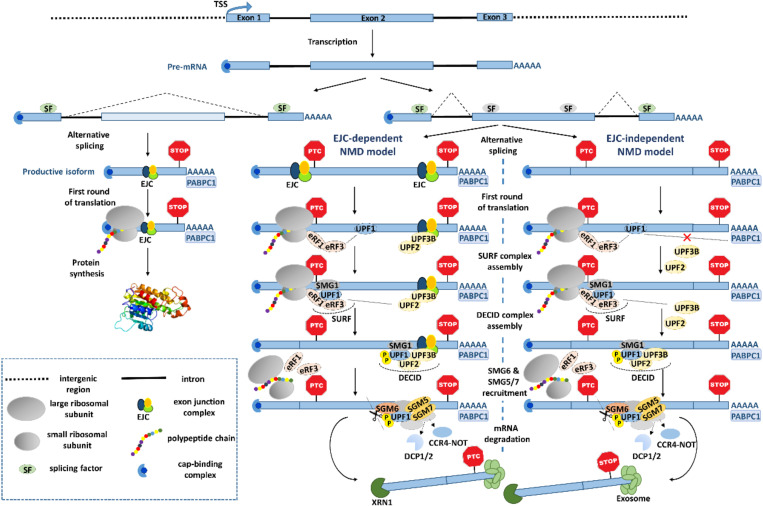
Figure 1.
Representation of the exon junction complex (EJC)-dependent and EJC-independent nonsense-mediated RNA decay (NMD) pathway, targeting a transcript generated by alternative splicing (AS). Introns are represented as black lines and exons as blue boxes. A gene is transcribed into a pre-mRNA and AS gives rise to two different mRNA isoforms. The left one represents a productive isoform that encodes a functional protein, and the two isoforms on the right include a premature stop codon (PTC)-containing exon that triggers NMD. The EJC-dependent model: During the first round of translation, the ribosome encounters a stop codon located more than 50-55 nucleotides upstream of an exon-exon junction, and eRF1/3 interact with UPF1 and SMG1, resulting in the SURF complex. Then, UPF1 interacts with UPF2 at the EJC, which ends in the DECID complex formation and activation of UPF1 through phosphorylation by SMG1. The EJC-independent model: If the interaction between PABPC1 and eRF3 is inhibited when the ribosome reaches a PTC, eRF3 can interact with UPF1 originating the SURF complex. Then, UPF2 and UPF3B diffused in the cytoplasm interact with UPF1, triggering UPF1 phosphorylation by SMG1 and the DECID complex assembly. The next steps are common to both models: The active UPF1 leads to its helicase function, rearranging the transcript to allow the recruitment of SMG5, SMG6, and SMG7. SMG6 cleaves the mRNA near the PTC, which results in unprotected ends. Meanwhile, SMG5 and SMG7 bind as a heterodimer to recruit the DCP1 and DCP2 decapping enzymes and the CCR4-NOT deadenylation complex. These RNA modifications allow 5′-3′ and 3′-5′ degradation by XRN1 and the exosome, respectively. TSS: Transcription start site.