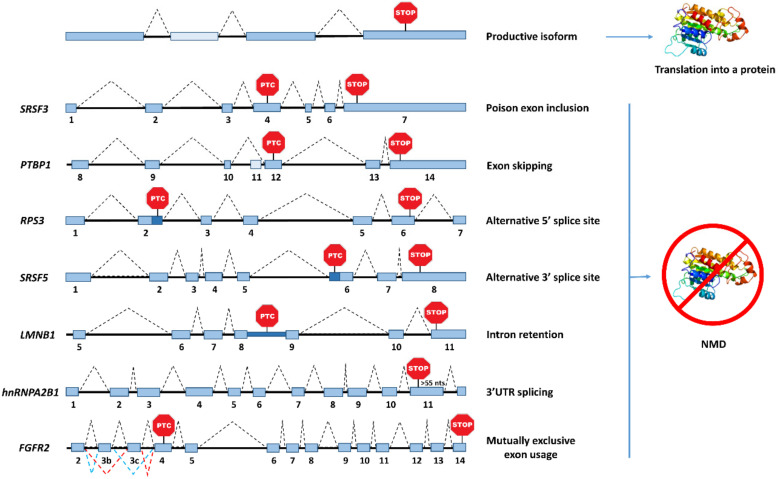
Figure 2.
Alternative splicing (AS) patterns inducing inclusion of a premature termination codon (PTC). Introns are represented as black lines and exons as blue boxes. The first splicing pattern listed in the image creates a full-length productive isoform that encodes a functional protein. The other represented transcripts contain a PTC that commits the isoform to nonsense-mediated mRNA decay (NMD). Cassette exon events, such as poison exon inclusion, induce retention of a PTC-containing exon, as shown for SRSF3 [78]. Exon skipping, another cassette exon event, may result in a frameshift leading to a PTC-positive isoform, as reported for PTBP1 [79]. Usage of an alternative 5′ or 3′ splice site, as well as intron retention, can include a segment of RNA with an in-frame PTC, turning the transcript into an NMD target, as represented above for RPS3, SRSF5, and LMNB1, respectively [71,80,81,82]. Splicing in the 3′ untranslated region (UTR) of hnRNPA2B1 can recruit an exon junction complex to a position located more than 55 nucleotides downstream of the stop codon, creating a premature context that triggers NMD [72]. The last example represents the inclusion of two exons in FGFR2, which are mutually exclusive, but if both exons are included or neither exon are in the splice variant, they introduce a frameshift that creates a PTC [83].