Figure 1.
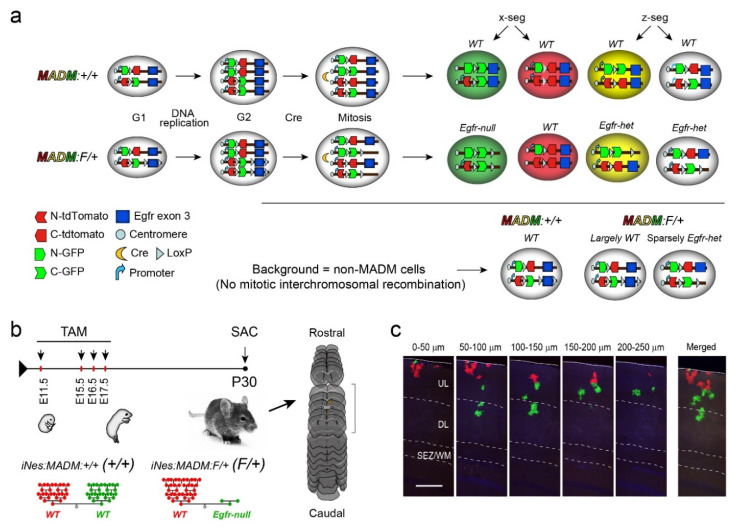
Clonal Analysis of Gliogenesis using Mosaic Analysis with Double Markers (MADM). (a) Molecular basis of labeling individual progenitor clones with distinct fluorescent reporters (GFP, green and tdTomato, red). Cre mediated interchromosomal mitotic recombination is followed by two distinct segregation patterns (x-seg and z-seg) generating clonal siblings in distinct colors as indicated. In control MADM mice, all cells will be WT and referred to as +/+. When a single floxed allele for Egfr is introduced, green cells lack Egfr (Egfr-null), while the red cells are maintained as WT (F/+ mice). Background genotypes obtained from the described genetic combinations using a tamoxifen induced Nestin-creERT2 transgene result in substantial differences in the Egfr genotype in background cells (non-MADM recombination events) which exert non-autonomous effects on MADM cells. (b) Schematic of tamoxifen (TAM) induction time points during embryonic development, and processing of forebrains in serial sections. Inductions were performed in iNes:MADM:Egfr +/+ (+/+) and iNes:MADM:Egfr F/+ (F/+) mice, which result in the generation of clones containing green and red cells that become distinguished following TAM inductions. Green MADM F/+ glia (Egfr-null) fail to develop, while glial production is elevated in the red sibling’s population. (c) Confocal images of a rare symmetric “G” cortical clone in an E16.5 induced +/+ cortex. SEZ/WM, subependymal zone/white matter; DL, deep layers; UL, upper layers. Scale bar, 100 µm.