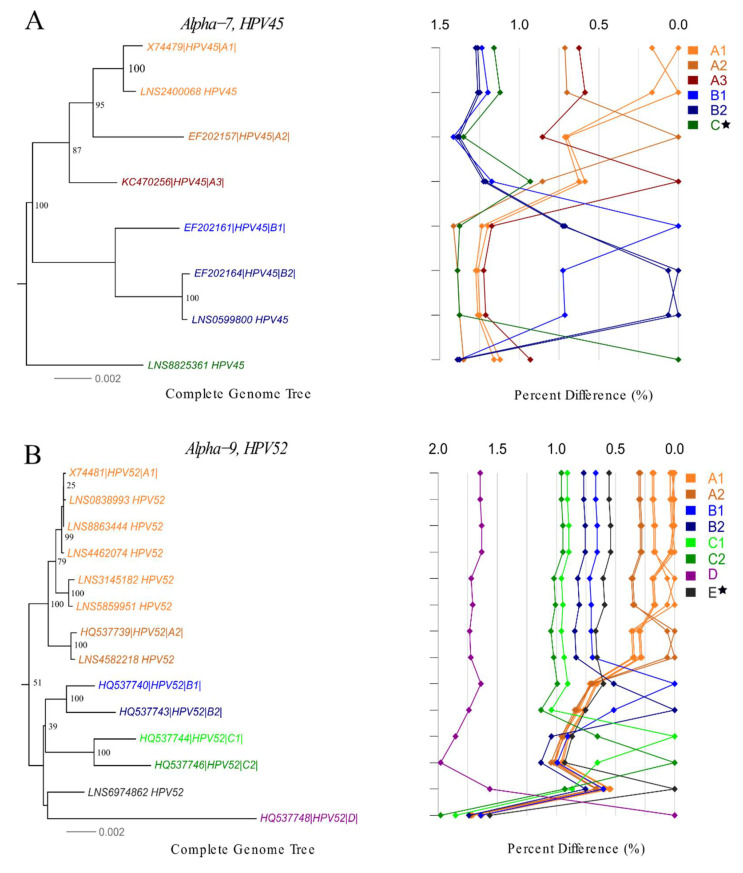
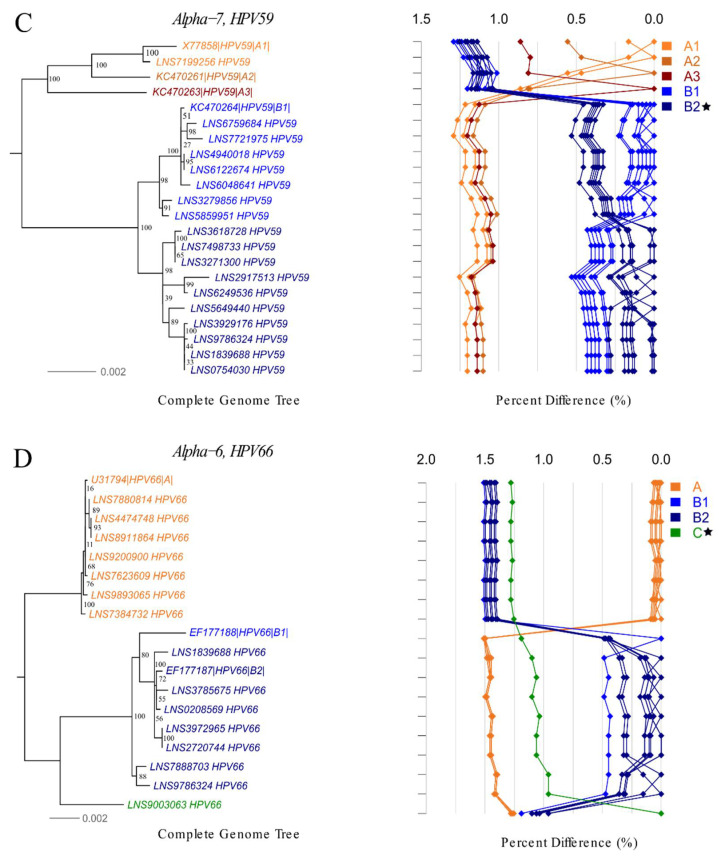
Figure 3.
Phylogenetic trees and pairwise distance comparisons for novel lineages/sublineages of high-risk HPV complete genome variants. Phylogenetic trees were inferred from a global alignment of the complete genome nucleotide sequences of the variants from following HPV genotypes using RAxML: (A) HPV45, (B) HPV52, (C) HPV59, (D) HPV66. A star ( ) indicates novel lineage/sublineage. The phylogenetic topology is shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.
) indicates novel lineage/sublineage. The phylogenetic topology is shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.