Abstract
Breast cancer is the most common cancer in women in the world. The role of germline genetic variability in ATP-binding cassette (ABC) transporters in cancer chemoresistance and prognosis still needs to be elucidated. We used next-generation sequencing to assess associations of germline variants in coding and regulatory sequences of all human ABC genes with response of the patients to the neoadjuvant cytotoxic chemotherapy and disease-free survival (n = 105). A total of 43 prioritized variants associating with response or survival in the above testing phase were then analyzed by allelic discrimination in the large validation set (n = 802). Variants in ABCA4, ABCA9, ABCA12, ABCB5, ABCC5, ABCC8, ABCC11, and ABCD4 associated with response and variants in ABCA7, ABCA13, ABCC4, and ABCG8 with survival of the patients. No association passed a false discovery rate test, however, the rs17822931 (Gly180Arg) in ABCC11, associating with response, and the synonymous rs17548783 in ABCA13 (survival) have a strong support in the literature and are, thus, interesting for further research. Although replicated associations have not reached robust statistical significance, the role of ABC transporters in breast cancer should not be ruled out. Future research and careful validation of findings will be essential for assessment of genetic variation which was not in the focus of this study, e.g., non-coding sequences, copy numbers, and structural variations together with somatic mutations.
Keywords: ABC transporter, therapy response, disease-free survival, breast cancer, next-generation sequencing, competitive allele-specific PCR
1. Introduction
Breast cancer (Online Mendelian Inheritance in Man, OMIM no. 114480) is the most common cancer in women in the world [1]. One of the frequently studied reasons for the lack of successful treatment outcomes in a considerable portion of the patients is multidrug resistance [2]. Multidrug resistance can be caused by ATP-binding cassette (ABC) transporters, e.g., by higher efflux of drugs out of tumor cells by P-glycoprotein (Multidrug resistance, MDR coded by ABCB1 gene) [2,3].
ABC transporters represent a large superfamily of membrane transporter proteins classified into seven families and translocate numerous compounds across intra and extracellular membranes. Their substrates include metabolic products, sterols, lipids, and xenobiotics [3]. However, of the total number of 48 active human ABC transporters, up to only 16 are able to transport anticancer drugs [3]. Most of the drug resistance is ascribed to largely studied multidrug resistance-related transporters ABCB1 (MDR, OMIM no. 171050), ABCC1 (MRP, OMIM no. 158343), and ABCG2 (BCRP, OMIM no. 603756) [4]. Proteins of the family ABCA are mostly lipid sterol transporters and can be associated with several diseases, e.g., Tangier or Alzheimer disease [5]. Their roles in cancer progression and the metastatic potential linked to lipid trafficking recently became the focus of numerous studies [6]. ABCBs participate in antigen processing and immune deficiency. Apart from ABCB1, a promiscuous and ubiquitous efflux pump [2], ABCB members also represent transporters of heme and bile acids [7]. Family C is mostly dedicated to multidrug resistance (MRP1-6) [8,9], but ABCC6, ABCC7, and ABCC8/9 are linked to serious diseases (pseudoxanthoma elasticum, cystic fibrosis, and diabetes mellitus, respectively) [9]. ABCDs are responsible for transport of fatty acids from peroxisomes to the cytoplasm [10] and ABCGs transport various products of metabolism, xenobiotics including anti-cancer drugs, bile acids, and steroids [11]. The rest of the transporters are not involved in transport, but rather act as translational inhibitors or protein synthesis regulators (ABCFs and ABCEs) [12].
Our recent pharmacogenomics study revealed a prognostic and predictive potential in a number of alterations in breast cancer [13]. The studied genes were implicated in the metabolism and transport of drugs administered to breast cancer patients in the clinics, clearly documenting the importance of this tool for the personalized medicine. The study provided a proof-of-the principle for the design and bioinformatics methodology, namely, the assembly and testing of an in-house pipeline for variant prioritization. Given the total number of 509 genes screened by the next generation sequencing (NGS), only a portion of variants could be validated in a subsequent phase. In order to select the most relevant functional alterations from the statistically significant set of variants, we down-sampled the results using information from in silico predictions and according to previously confirmed pharmacogenomic and clinical evidence. Thus, some potentially useful biomarkers of prognosis or prediction of therapy outcome could have been missed.
In the present study, we aimed to use less strict criteria for investigating the importance of germline genetic variability in coding, untranslated regions (UTR), and adjacent regions of all human members of the ABC superfamily for prognosis and response to cytotoxic therapy of breast cancer patients. All variants in ABCs identified in the testing phase were correlated with disease-free survival (DFS) and response of the patients to preoperative cytotoxic therapy, and thoroughly reviewed, including permutation analysis, evaluation of haplotypes, and gene dosage. We have not addressed functional relevance to enable identification of purely correlative biomarkers. Prioritized variants were further validated in a large cohort of breast cancer patients from a single population. Thus, the present study brings a more detailed view of the relevance of genetic variability of ABC transporters for breast cancer prognosis and therapy outcome predictions.
2. Results
2.1. Testing Phase
The clinical characteristics of the patients (n = 105) are in Table S1. The subgroup of patients was treated with the neoadjuvant cytotoxic therapy (NACT) (n = 68) and the response to this treatment was available. The rest of patients were treated with adjuvant cytotoxic therapy based on monotherapy or combinations of anthracyclines, cyclophosphamide, 5-fluorouracil, and taxanes. DFS was evaluated for all patients and the mean follow-up of the patients was 70 ± 28 months.
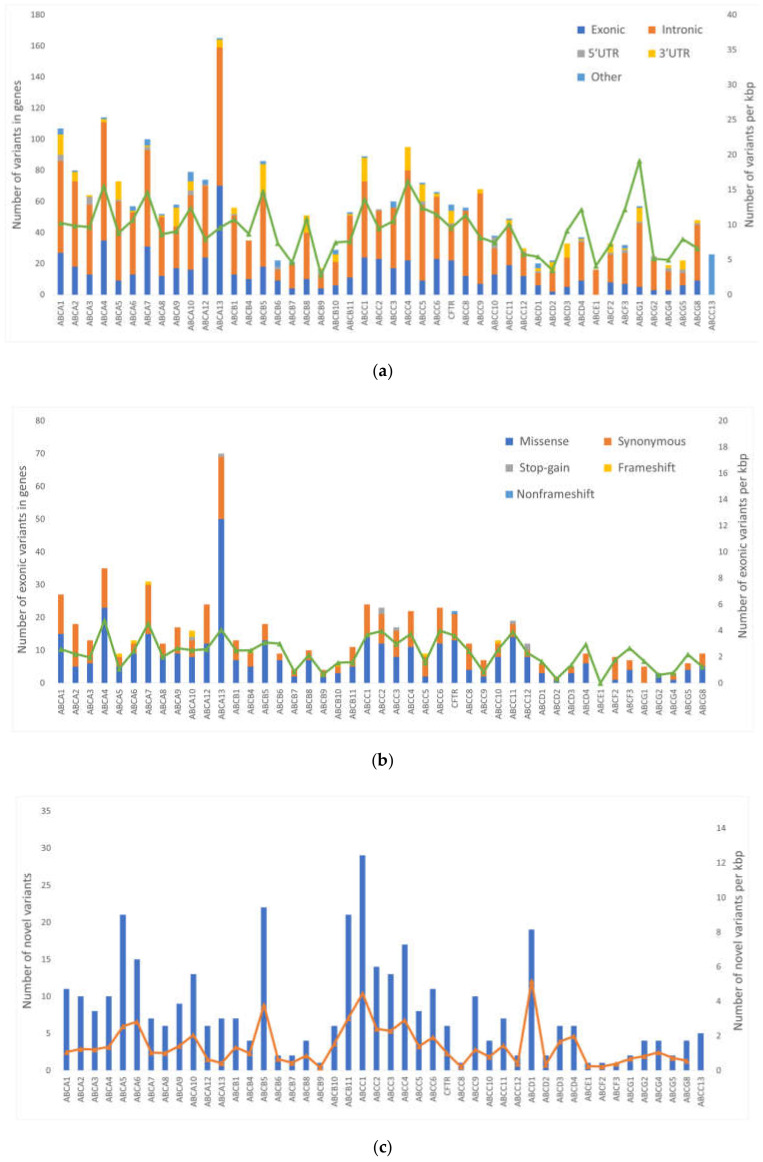
The average coverage was 82.3 ± 29.1 and 95% of the captured regions were covered at least ten times. Altogether, we found 2611 variants in exonic and adjacent intronic sequences. Of the total number of 48 genes and one pseudogene (ABCC13) subjected to analysis, 46 genes (94%) contained at least one genetic alteration. No alterations were found in ABCF1, TAP1 (alias ABCB2), and TAP2 (ABCB3) genes. On the other hand, the most polymorphic genes, with over one hundred alterations, were ABCA13 (165 alterations), ABCA4 (114), and ABCA1 (109). Of the total number of 2611 variants, 636 were in exons, 1544 intronic, and 253 were in 3’UTR or 5’UTR regions according to National Center for Biotechnology Information (NCBI) Reference Sequence Database (RefSeq; https://www.ncbi.nlm.nih.gov/refseq/) in Annovar (Table 1).
Table 1.
Observed alterations in ATP-binding cassette (ABC) transporters divided by function according to Annovar.
| Type | Total | Percentage |
|---|---|---|
| Intronic | 1544 | 59.1 |
| Exonic (coding) | 636 | 24.4 |
| UTR’3 | 204 | 7.8 |
| Intergenic | 76 | 2.9 |
| UTR’5 | 49 | 1.9 |
| Downstream 1 | 28 | 1.1 |
| Upstream 1 | 26 | 1.0 |
| Splicing 2 | 13 | 0.5 |
| Other 3 | 35 | 1.3 |
1 Variants are 1 kb from transcription end/start site; 2 Variants are 2 bp within splice junction. 3 Exonic/intronic non-coding RNA, or variant in overlapping regions (upstream–downstream) of two different genes.
On average, each patient showed 608 ± 33 variants. We found 17 loss-of-function (LOF) truncating variants that were either affecting the stop codon (stop-gain) or frameshift insertions or deletions (indels). There were 355 of the variants that were non-synonymous single nucleotide variants (SNVs) and 263 that were synonymous SNVs (Table 2). In total, 1058 variants (41%) had minor allele frequency (MAF) > 0.05, and the rest of the 1553 variants, had MAF ≤ 0.05.
Table 2.
Overview of the observed exonic alterations in ABC transporters by coding consequence.
| Classification | Total | Percentage |
|---|---|---|
| Non-synonymous SNV | 355 | 55.8 |
| Synonymous SNV | 263 | 41.4 |
| Stop-gain | 8 | 1.3 |
| Frameshift deletion | 6 | 0.9 |
| Frameshift insertion | 3 | 0.5 |
| Non-frameshift deletion | 1 | 0.1 |
Altogether, 256 (10%) of the variants were novel (i.e., not found in dbSNP Build 150). Out of these, 162 had MAF > 0.01. The distribution of the variants according to their functional classes and frequencies of novel variants in the groups of genes are shown in Figure 1.
Figure 1.
Distribution of alterations in individual ABC transporter genes. The picture shows: (a) the frequency of genetic alterations according to their functional classes; (b) the frequency of genetic alterations according to their exonic functional classes analyzed by RefSeq: National Center for Biotechnology Information (NCBI) Reference Sequence Database (https://www.ncbi.nlm.nih.gov/refseq/) shown according to individual transporters (excluding ABCC13 pseudogene); and (c) the number of novel variants according to individual transporters. The number of the variants normalized to the transcript length in kilo base pairs (kbp) per each gene are shown for each plot on the right axis and depicted by the overlaid line.
Variants departing from Hardy–Weinberg equilibrium (p < 0.01, n = 101) were excluded from analyses and further only variants with MAF > 0.05 were considered relevant to achieve adequate statistical power in the validation phase. In addition, variants with the missing data in more than 50% of patients were excluded (n = 54). Application of these filtration criteria resulted in a set of 903 variants which were further evaluated for associations with the response of patients to NACT and DFS. We found 56 variants associated with the response to NACT and 47 variants associating with DFS. Six variants reported significant in the previous study [13] were further excluded. Following haplotype evaluations, 38 variants were considered tagged (>80%) with some other variant and were not assessed further. As a result, 22 variants associated with the response and 37 variants associated with DFS were followed. The gene dosage relationship was then evaluated for variants associating with DFS and variants not fulfilling this condition were excluded (n = 7). Neither of these variants was significant in the recessive genetic model (variant allele versus wild type). Following these control steps, 52 variants (45 SNVs and 7 indels, Table S2) were prioritized for the validation phase in a larger cohort of breast cancer patients.
2.2. Validation Phase
The clinical characteristics of the patients (n = 802) are summarized in Table S3. A subgroup of patients treated with NACT composed of 168 patients. In total, 371 patients received adjuvant cytotoxic therapy. Patients with localized disease and good prognosis were untreated (n = 83) and a portion of patients was treated only with hormonal therapy (n = 311). The mean follow-up of the patients was 76 ± 30 months.
Out of 52 variants subjected to genotyping, attempts to optimize detection techniques failed in 10 variants (5 indels and 5 SNVs) and could not be further evaluated for clinical associations. One variant was additionally included into the list (rs2893007) based on haplotype tagging (r2 = 1) to replace the variant rs11764054 whose analysis failed. No tagging variants (r2 > 0.8) were found to replace the rest of the failed variants. In the end, 43 variants were successfully genotyped. No variants significantly departed from Hardy–Weinberg equilibrium and less than 1% of theoretical data points were missing due to uncertainty in genotype calling, or an absent signal. MAFs of variants in the validation phase did not substantially differ from those observed in the testing one (Table 3).
Table 3.
Distribution of genotypes for variants assessed in the validation phase.
| Gene | SNP ID 1 | Genotype Distribution 2 | MAF 3 | |||
|---|---|---|---|---|---|---|
| Common Homozygotes | Heterozygotes | Rare Homozygotes | Validation Set | Testing Set | ||
| ABCA1 | rs41474449 | 658 | 136 | 3 | 0.09 | 0.07 |
| ABCA4 | rs537831 | 377 | 342 | 78 | 0.31 | 0.31 |
| ABCA4 | rs2065711 | 436 | 309 | 55 | 0.26 | 0.20 |
| ABCA4 | rs2275032 | 540 | 230 | 30 | 0.18 | 0.14 |
| ABCA4 | rs2275033 | 270 | 396 | 133 | 0.41 | 0.40 |
| ABCA4 | rs3789398 | 353 | 361 | 84 | 0.33 | 0.35 |
| ABCA5 | rs1420904 | 679 | 113 | 7 | 0.08 | 0.08 |
| ABCA5 | rs2067851 | 681 | 112 | 2 | 0.07 | 0.07 |
| ABCA7 | rs9282562 | 604 | 188 | 8 | 0.13 | 0.14 |
| ABCA8 | rs4147976 | 318 | 358 | 121 | 0.38 | 0.35 |
| ABCA9 | rs2302294 | 368 | 352 | 80 | 0.32 | 0.34 |
| ABCA9 | rs11871944 | 326 | 366 | 103 | 0.36 | 0.40 |
| ABCA12 | rs71428357 | 726 | 70 | 3 | 0.05 | 0.08 |
| ABCA13 | rs7780299 | 597 | 187 | 16 | 0.14 | 0.12 |
| ABCA13 | rs17132289 | 687 | 106 | 6 | 0.07 | 0.08 |
| ABCA13 | rs17548783 | 201 | 400 | 192 | 0.49 | 0.49 |
| ABCA13 | rs28637820 | 628 | 163 | 8 | 0.11 | 0.13 |
| ABCA13 | rs74859514 | 665 | 124 | 10 | 0.09 | 0.08 |
| ABCB1 | rs9282564 | 609 | 168 | 21 | 0.13 | 0.13 |
| ABCB5 | rs3210441 | 283 | 400 | 116 | 0.40 | 0.44 |
| ABCB5 | rs12700230 | 466 | 285 | 49 | 0.24 | 0.23 |
| ABCB5 | rs2893007 | 676 | 120 | 5 | 0.08 | 0.10 |
| ABCB8 | rs2303922 | 336 | 362 | 100 | 0.35 | 0.34 |
| ABCB11 | rs853772 | 203 | 403 | 190 | 0.49 | 0.25 |
| ABCC1 | rs4148379 | 456 | 287 | 48 | 0.24 | 0.20 |
| ABCC2 | rs2273697 | 478 | 273 | 39 | 0.22 | 0.22 |
| ABCC3 | rs8077268 | 649 | 147 | 4 | 0.10 | 0.10 |
| ABCC3 | rs12604031 | 271 | 374 | 154 | 0.43 | 0.44 |
| ABCC4 | rs899494 | 583 | 198 | 17 | 0.15 | 0.12 |
| ABCC4 | rs2274405 | 339 | 352 | 102 | 0.35 | 0.37 |
| ABCC5 | rs4148579 | 259 | 404 | 137 | 0.42 | 0.43 |
| ABCC5 | rs12638017 | 686 | 111 | 3 | 0.07 | 0.06 |
| ABCC8 | rs739689 | 349 | 356 | 91 | 0.34 | 0.40 |
| ABCC10 | rs75320251 | 654 | 135 | 8 | 0.09 | 0.09 |
| ABCC11 | rs17822931 | 592 | 184 | 21 | 0.14 | 0.14 |
| ABCC13 | rs2254297 | 254 | 381 | 160 | 0.44 | 0.40 |
| ABCC13 | rs2822582 | 306 | 369 | 121 | 0.38 | 0.40 |
| ABCD4 | rs2301346 | 394 | 334 | 67 | 0.29 | 0.32 |
| ABCD4 | rs2301347 | 305 | 376 | 120 | 0.38 | 0.40 |
| ABCF2 | rs79537035 | 527 | 242 | 30 | 0.19 | 0.23 |
| ABCG8 | rs34198326 | 685 | 109 | 4 | 0.07 | 0.06 |
| ABCG8 | rs56260466 | 685 | 104 | 9 | 0.08 | 0.06 |
| CFTR | rs34855237 | 538 | 229 | 30 | 0.18 | 0.08 |
1 Reference number in dbSNP (https://www.ncbi.nlm.nih.gov/snp/); 2 Genotypes do not sum up to 802 due to missing data; 3 MAF = minor allele frequency.
We further evaluated associations of variants with the response and DFS of patients in the validation set. For six variants with less frequent homozygous genotypes (n < 5), the recessive genetic model was used for the sake of the statistical power of comparisons. The variants that associated with response in both testing and validation phases are listed in Table 4 and thus these variants are considered replicated with putative clinical importance. However, none of these associations passed the false discovery rate (FDR) test for multiple testing (q = 0.0025) and, thus, cannot be deemed statistically significant after such correction.
Table 4.
Validated variants in ABC transporters significantly associating with the response of patients to the neoadjuvant cytotoxic therapy.
| Gene | SNP ID | Genotype | Good Response 1 | Poor Response 1 | χ-Square | p-Value |
|---|---|---|---|---|---|---|
| ABCA4 | rs2275032 | AA | 75 | 33 | 6.33 | 0.042 |
| CA | 48 | 7 | ||||
| CC | 4 | 1 | ||||
| ABCA9 | rs11871944 | CC | 48 | 24 | 6.76 | 0.034 |
| CT | 61 | 14 | ||||
| TT | 18 | 2 | ||||
| ABCA12 | rs71428357 | CC | 103 | 41 | 8.32 | 0.004 |
| CT + TT | 22 | 0 | ||||
| ABCB5 | rs3210441 | GG | 42 | 11 | 6.22 | 0.045 |
| GA | 62 | 28 | ||||
| AA | 23 | 2 | ||||
| ABCC5 | rs4148579 | CC | 43 | 6 | 8.55 | 0.014 |
| CT | 68 | 24 | ||||
| TT | 15 | 11 | ||||
| ABCC8 | rs739689 | AA | 55 | 11 | 6.81 | 0.033 |
| GA | 60 | 21 | ||||
| GG | 11 | 9 | ||||
| ABCC11 | rs17822931 | CC | 89 | 37 | 6.42 | 0.011 |
| CT + TT | 37 | 4 | ||||
| ABCD4 | rs2301347 | CC | 53 | 9 | 8.59 | 0.014 |
| CA | 60 | 21 | ||||
| AA | 14 | 11 | ||||
| ABCD4 | rs2301346 | TT | 73 | 15 | 7.28 | 0.026 |
| CT | 45 | 20 | ||||
| CC | 7 | 6 |
1 Numbers of patients with specified genotypes divided by response to the neoadjuvant treatment.
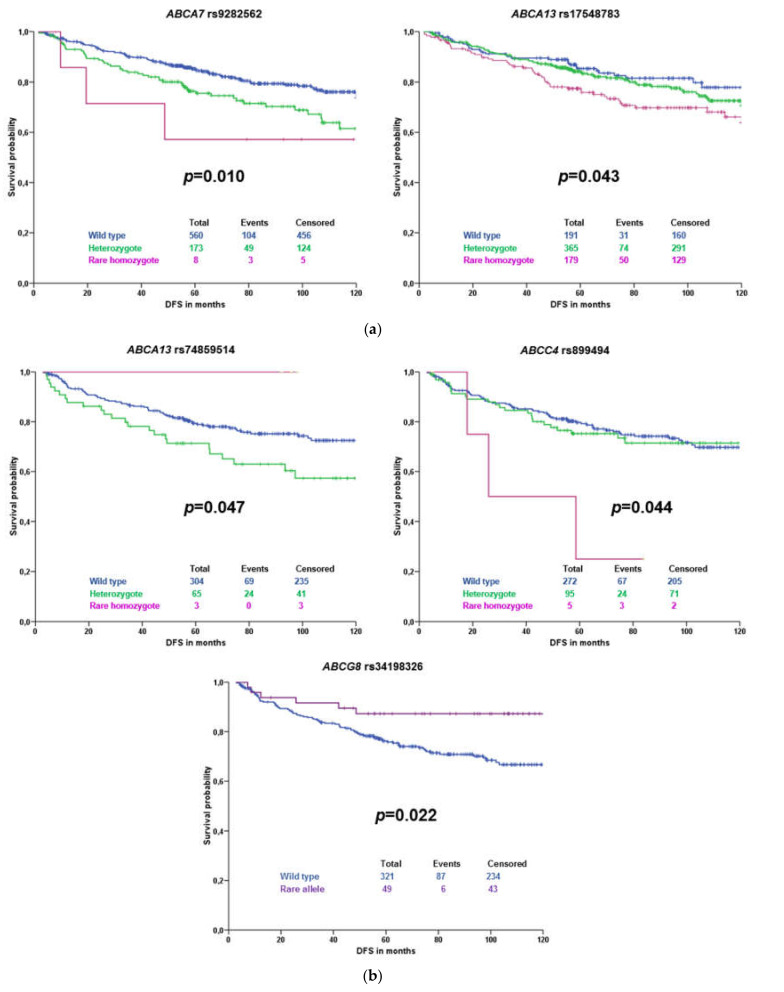
Subsequently, associations of variants with DFS of all patients and patients stratified according to the received therapy were evaluated. Significant results for all patients with complete follow-up data (n = 744) are displayed in Figure 2a. Results for patients treated with cytotoxic therapy (n = 371) are presented in Figure 2b. No significant association was observed in a subgroup of patients treated only with hormonal therapy (n = 312).
Figure 2.
Kaplan–Meier plots with validated associations of variants with disease-free survival (DFS) of all patients unstratified (a) or subgroup of patients treated with cytotoxic therapy (b). The blue line represents the common homozygous genotype, the green line the heterozygote, and the magenta line the rare homozygote. The violet color represents rare variant carriers. Significance was evaluated by the log-rank test; numbers represent individuals in compared groups.
Of these variants, rs74859514 did not pass the gene dosage condition (Figure 2b). None of the associations has passed the FDR test for multiple testing (q = 0.0023) and, thus, cannot be further considered statistically validated.
We further clarified the effect of molecular subtype on prognosis of the patients by their stratification according to the intrinsic molecular subtype. Associations with DFS were then calculated separately for each subtype (Table 5 and Figure S1).
Table 5.
Validated associations of variants in ABC transporters associating with DFS of patients treated with cytotoxic therapy according to their molecular subtypes.
| Gene | SNP ID | Genotypes | Subtypes 1 | |||
|---|---|---|---|---|---|---|
| Luminal A | Luminal B | HER2 | TNBC | |||
| All patients (n = 744) | ||||||
| ABCA7 | rs9282562 | Common homozygous | 165 | 206 | 39 | 63 |
| Rare allele | 45 | 68 | 17 | 21 | ||
| p-value 2 | 0.626 | 0.316 | 0.010 | 0.325 | ||
| ABCA13 | rs17548783 | Common homozygous | 58 | 67 | 12 | 24 |
| Heterozygous | 112 | 129 | 31 | 40 | ||
| Rare homozygous | 39 | 74 | 13 | 20 | ||
| p-value 2 | 0.050 | 0.114 | 0.492 | 0.039 | ||
| Patients treated with cytotoxic therapy (n = 371) | ||||||
| ABCA13 | rs74859514 | Common homozygous | 62 | 125 | 25 | 58 |
| Rare allele | 12 | 27 | 11 | 8 | ||
| p-value 2 | 0.441 | 0.606 | 0.001 | 0.009 | ||
| ABCC4 | rs899494 | Common homozygous | 50 | 114 | 27 | 46 |
| Rare allele | 24 | 38 | 9 | 20 | ||
| p-value 2 | 0.825 | 0.415 | 0.050 | 0.565 | ||
| ABCG8 | rs34198326 | Common homozygous | 63 | 135 | 30 | 54 |
| Rare allele | 11 | 17 | 5 | 11 | ||
| p-value 2 | 0.094 | 0.040 | 0.847 | 0.091 | ||
1 Numbers of patients for each genotype stratified by molecular subtypes are displayed; HER2 = HER2 enriched, TNBC = triple negative breast cancer. 2 p-value departed from log-rank test (significant results in bold).
This analysis showed that prognostic associations differ according to the molecular subtype. In the whole group of patients, rs9282562 in ABCA7 and rs17548783 in ABCA13 were prognostic only for HER2 enriched and triple negative (TNBC) subtypes, respectively. In the subgroup treated with cytotoxic therapy, again single nucleotide polymorphisms (SNPs) were prognostic for patients with HER2 enriched and TNBC subtypes (rs74859514 in ABCA13). Carriers of the rare variant in ABCG8 rs34198326 had better DFS than those with the common homozygous genotype—this association was significant only in patients with the luminal B subtype.
In order to clarify associations of variants with gene expression, we used transcriptomic data from previous gene expression profiling [14] and compared it with variants that significantly associated with DFS or response to NACT in the validation set (n = 168 patients for whom gene expression was available). We also analyzed expression quantitative trait loci (eQTL) associations of these variants using gene expression in healthy tissues available on the GTEx portal (https://www.gtexportal.org). The SNP rs17548783 was significantly associated with ABCA13 transcript levels in tumors assessed in the previous study [14] (Table 6), but no eQTL were found in the GTEx database. No eQTL were found also for rs2275032 in ABCA4, rs71428357 in ABCA12, rs3210441 in ABCB5, and rs34198326 in ABCG8. Significant results from eQTL analysis are summarized in Table 7 and Figure S2.
Table 6.
Association of single nucleotide polymorphism (SNP) rs17548783 in ABCA13 transporter with intratumoral transcript levels.
| SNP ID | Genotype | n | Expression 1 | S.D. 2 | p-Value |
|---|---|---|---|---|---|
| rs17548783 | Common homozygous | 9 | −7.29 | 2.11 | 0.015 |
| Heterozygous | 18 | −9.90 | 2.67 | - | |
| Rare homozygous | 7 | −9.45 | 2.67 | - |
1 Transcript levels expressed as Ct (cycle threshold) value of ABCA13 subtracted from mean Ct of reference genes [14]. 2 S.D. = Standard deviation.
Table 7.
eQTL analysis of SNPs significantly associating with patients’ DFS or response to neoadjuvant cytotoxic therapy (NACT).
| SNP ID | Gene | Tissue | Normalized Expression Trend | p-Value 1 |
|---|---|---|---|---|
| rs11871944 | ABCA9 | multiple 2 | CC > TC > TT | 3.1 × 10−7 |
| rs4148579 | ABCC5 | multiple 2 | CC > TC > TT3 | 3.5 × 10−33 |
| rs739689 | ABCC8 | brain (cerebellum) | AA > AG > GG | 8.6 × 10−9 |
| rs17822931 | ABCC11 | brain | CC > CT > TT | 6.9 × 10−5 |
| LONP2 4 | breast | CC > CT > TT | 1.4 × 10 −4 | |
| rs2301347 | ABCD4 | multiple 2 | AA > CA > CC | 5.0 × 10−20 |
| lnc-SYNDIG1L-2 4 | breast | AA > CA > CC | 1.8 × 10−16 | |
| rs2301346 | ABCD4 | multiple 2 | CC > TC > TT | 1.8 × 10−12 |
| lnc-SYNDIG1L-2 4 | breast | CC > TC > TT | 4.2 × 10−15 | |
| rs9282562 | ABCA7 | multiple 2 | ref > het > delTG | 1.3 × 10−11 |
| s74859514 | UPP1 4 | cerebellum, muscle | GG > GC > CC | 2.7 × 10−5 |
| rs899494 | ABCC4 | thyroid, whole blood | AA > AG > GG | 1.9 × 10−16 |
1p-value of the most significant association is shown. 2 Significant results in more than three different tissues. 3 The highest expression is seen for TT genotype in whole blood and esophageal mucosa; the opposite i.e., highest expression of CC genotype is seen for the rest of the tissues. 4 Alternative eQTL.
3. Discussion
The role of germline genetic variability among ABC transporters in prognosis of breast cancer patients as well as in their response to chemotherapy is underexplored. In our previous publication, we dealt with pharmacologically relevant germline genetic polymorphisms in 509 breast cancer-related genes [13]. In the present study, we used the same approach to reveal all associations of genetic variants in human ABC transporters with chemotherapy response and survival of the patients.
A total of 2611 variants were found in a testing set. The majority of variants were found in intronic regions. Lower numbers of variants were found in coding regions and UTRs. Interestingly, no variants were found in ABCF1, TAP1 (alias ABCB2), and TAP2 (ABCB3). TAP1 and TAP2 are antigen presenting transporters and alterations in these genes associate with autoimmune diseases, susceptibility to infections, or malignancies [15]. Similarly, ABCF1 plays a role in the regulation of inflammatory processes [16] and alterations in ABCF1 are linked with autoimmune diseases as well [17]. Therefore, it seems that genetic variants in these genes negatively impacts immunity and inflammatory processes which explain limited variability, in line with our findings. On the other hand, the most variable genes were ABCA13 (165 alterations), ABCA4 (114), and ABCA1 (109). The members of ABCA family are typically large genes (transcript length 7‒17 kbp) and thus likely to accumulate variants. When normalized for the length of transcript, ABCG1, ABCC4, and ABCA4 have the highest count of variants per kbp, ranging from 16 to 20. Interestingly, ABCA4, ABCA7, and ABCA13 had the highest variant counts in exonic regions (4.1‒4.8 variants per kbp). We found several LOF variants in ABC transporters—eight stop-gains and nine frameshift indels. These events have high impact on function of the protein. Moreover, all 17 LOF variants were present in genes of the first quartile of the most intolerant genes to LOF events [18]. These facts advocate for further investigation of LOF variants in ABC transporters. Unfortunately, LOF variants are rare. For the sake of maintaining enough statistical power for comparison with clinical data, only common variants (MAF > 0.05) could be used in the present study.
In total, we selected 903 variants and subjected them to a thorough statistical analyses. Of these variants, 43 associated with response or DFS and were capable of validation in a cohort of 802 breast cancer patients. Five associations with DFS and nine with response to NACT were replicated in the validation set. If multiple simultaneous statistical tests are calculated, a type I error (a risk of false positive results) occurs. To prevent this error, correction for multiple testing must be used. There are several methods to do so. Here, we applied the wildly used FDR, a test by Benjamini–Hochberg. After this correction, none of the associations of variants with clinical features remained significant and, thus, cannot be considered validated. Nevertheless, we found some interesting associations which we will discuss further.
ABCA13 is responsible for lipid transport and variants in this gene can cause schizophrenia [5]. Carriage of the rare allele of SNP rs17548783, located in downstream intronic region of ABCA13, was associated with shorter DFS of patients in our study. Based on our findings, a rare allele of this variant, significantly associated with lower ABCA13 intratumoral transcript levels in a validation set (Table 6). Lower transcript levels of ABCA13 were associated with worse response to NACT in a previous study [14], further underpinning the role of this SNP as a putative poor prognosis biomarker. This consequence is the most interesting link observed at present. Nevertheless, the response to NACT does not significantly associate with DFS in our datasets a fact that clearly calls for further research.
Another variant in ABCA13, the missense rs74859514 (Ala2223Pro), associated with DFS in patients treated with chemotherapy, but without gene dosage relationship. Neither of these two SNPs has records in the present literature, although associations of ABCA13 with patients’ outcome have been described in several previous studies. A decreased expression of ABCA13 was associated with shorter DFS in 51 glioblastoma patients [19] and 51 colorectal cancer patients [20]. The opposite was found for ovarian cancer (n = 77) and higher levels of ABCA13 predicted worse overall survival in ovarian cancer patients [21]. Amplification of 7p12 (which includes ABCA13 and HUS1, EGFR, and IKZF1) predicted worse response to NACT in muscle-invasive bladder cancer [22]. Such contradictory results from different cancers are puzzling. Despite we must bear in mind that none of the associations found in our study passed the FDR test, some may still have clinical potential. Additional studies will be needed to confirm these results.
A synonymous variant rs71428357 in ABCA12 associated with response to NACT. Patients responding well to chemotherapy were more often carriers of the rare allele. Synonymous variants can affect RNA splicing, folding, and stability [23] and are associated with several diseases, such as Alzheimer disease, pulmonary sarcoidosis, galactosemia, or cancer [24]. The role of this particular ABCA12 variant in cancer or other diseases is still unknown. However, higher ABCA12 transcript levels in non-tumor tissues associated with worse response to NACT in breast cancer patients in our previous study [14]. The opposite, i.e., higher levels associating with residual disease, was found by Park et al. [25]. Interestingly, we previously identified this gene among candidate ABCs with predictive or prognostic potential for patients with breast, colorectal, and pancreatic carcinomas [26].
Among other members of the ABCA family, associations with response to NACT were observed for ABCA4 (variant rs2275032) and ABCA9 (rs11871944). A deletion in ABCA7 (rs9282562) associated with shorter DFS of the patients. These variants are not described in the present literature, however, higher transcript levels of ABCA9 significantly associated with worse survival in high-grade serous ovarian cancer tumors [6]. Silencing of ABCA7 reduces epithelial to mesenchymal transition in ovarian cancer cell lines and knockdown of ABCA7 inhibited migration, cell proliferation, and invasion [27]. In addition, lower ABCA7 levels associated with shorter DFS of colorectal cancer patients [20].
ABCB5 confers 5-fluorouracil resistance and promotes cell invasiveness in colorectal cancer [28]. Variant rs3210441 in ABCB5 associated with response to NACT in our study, but no eQTL was found and additional supportive data about the role of this SNP or protein in breast cancer is lacking.
Protein coded by ABCC11 is responsible for transport of bile acids, conjugated steroids, or cyclic nucleotides. Diseases linked with this gene include malfunction of apocrine gland secretion and lateral sinus thrombosis (https://www.genecards.org). ABCC11 is a transporter of 5-fluorouracil [3]. In our study, a missense ABCC11 variant rs17822931 (Gly180Arg) associated with response to NACT. Carriers of the wild-type allele had significantly poorer outcomes than patients with an alternative allele. This variant is known for its determination of human earwax type [29]. It is associated with breast cancer risk in the Japanese population [30]. This variant is also linked with axillary osmidrosis, colostrum secretion in the mammary gland, and mastopathies [31]. Wild type allele C also confers to chemotherapy resistance to 5-fluorouracil by exporting active metabolite 5-fluoro-2′-deoxyuridine 5′-monophosphate (FdUMP) [32]. ABCC11 expression (together with ABCB1) is responsible for resistant phenotype of breast cancer cell lines resistant to eribulin and inhibition of ABCC11 can partially restore the cross-resistance to 5-fluorouracil [33]. Higher ABCC11 gene expression was also associated with poor response to NACT in breast cancer patients [25]. Interestingly, this SNP is associated with expression of ABCC11 only in the brain, but with LONP2, coding mitochondrial matrix protein, in breast tissue (Table 7). Relations between mastopathy, breast cancer risk, and, after chemotherapy, even drug resistance suggest strong connection of this variant to breast cancer. Association with response to chemotherapy of breast cancer patients has been suggested previously [31], our result corroborates this assertion.
Among other members of the ABCC family, ABCC5 (rs4148579) and ABCC8 (rs739689) associated with response to NACT and ABCC4 (rs899494) with DFS of the patients. ABCC4 was among amplified genes in resistant cancer cell lines [34]. The ABCC4 gene was also identified to play a role in cellular migration of breast cancer cell line models MCF-7 and MDA-MB-231 [35]. In our previous study [14], we have seen associations of high ABCC8 transcript levels with low grade and negative/positive status of estrogen receptor. Additionally, the expression level non-significantly (p = 0.096) associated with worse responses of breast cancer patients to NACT [14]. Nevertheless, in the present study we did not find association of rs739689 (intronic A > G transition) with ABCC8 transcript levels. eQTL associations at the GTEx portal are ambiguous. The wild-type AA genotype has the highest expression of ABCC8 in cerebellum, but no significant association can be found in breast tissue. This SNP is also highly significantly associated with expression of NCR3LG1, KCNJ11, and SNORD14 genes with fragmentary and elusive data on association with breast cancer. From the data discussed above, it can be summarized that the present knowledge is incomplete and, thus, no clear picture can be presented.
Unlike other ABCD transporters, ABCD4 is not found in peroxisomes, but in lysosomes. It takes part in transport of cobalamin (vitamin B12) and mutations in this transporter cause inherited defects of intracellular cobalamin metabolism [10]. Low transcript levels of this gene were also associated with shorter DFS of colorectal cancer patients [20] and ABCD4 was among amplified genes in resistant cancer cell lines [34]. In our study, wild-type variants rs2301347 and rs2301346 (both intronic) associated with the good response to NACT. Wild-type genotypes of these two variants show lower transcript levels of long non-coding (lnc) RNA lnc-SYNDIG1L-2 overlapping ABCD4 in mammary tissue (Table 7) suggesting potential clinical relevance. However, the lack of association with ABCD4 transcript levels that we found in our dataset precludes any strict conclusions.
ABCG8 is a transporter of sterols from hepatocytes and enterocytes [36]. The rare allele of its SNP rs34198326 was associated with longer DFS of chemotherapy treated patients in our study. Expression of ABCG8 was downregulated in tumors of breast cancer patients compared to non-neoplastic control tissues [14], but the role of germline polymorphism is unclear.
The role of ABC transporters in cancer has been known for a long time. Multidrug resistance has been studied since 1970, when it was first mentioned [37]. The well-studied ABCB1 gene (MDR1) was discovered in 1974 by V. Ling, and nearly twenty years later, the discovery of ABCC1 and ABCG2 concerning drug resistance was reported [2]. Although associations of ABCB1 gene expression with breast cancer prognosis were reported repeatedly, evidence for the role of its genetic variability in response to treatment is elusive. A recent review demonstrated that three frequently studied polymorphisms in ABCB1 (rs1045642, rs1128503, and rs2032582) cannot be considered reliable predictors of response to chemotherapy in breast cancer patients [38]. Similarly, an association of ABCC1 expression with the survival of breast cancer patients was described [39]. However, only a few studies on genetic polymorphisms can be found. ABCC1 variants rs4148350, rs45511401, and rs246221 associated with the risk of febrile neutropenia in patients treated with 5-fluorouracil, epirubicin, and cyclophosphamide [40] and it was very recently discovered that ABCC1 variant burden is a strong predictor of DFS in breast cancer patients rather than the genotype attributed to individual variants [41]. ABCG2 transports several drugs used for breast cancer treatment. In a recent study on the TCGA cohort, ABCG2 transcript levels associated with a decreased progression-free survival, although, gene variants (either somatic or germline) influenced ABCG2 expression only moderately [42]. From the above-reviewed information, it can be summarized that despite numerous studies on drug transporters utilization for predicting therapy outcome, strong support is still missing. Other transporters, with rather physiological roles, are much less explored in oncology, and studies were largely dedicated to gene expression in contrast with less studied genetic variability.
In conclusion, genetic variability in ABC transporters might play a role in breast cancer prognosis and help with prediction of therapy outcome of the patients. Although no alterations observed by this study can be considered statistically validated, particularly associations of downstream variant affecting expression, rs17548783 in ABCA13 with DFS and variant rs17822931 (Gly180Arg) in ABCC11 with response to NACT attract attention because of their support in the literature. These are interesting candidates for future research. Furthermore, elucidations are needed to explore additional genetic component, e.g., non-coding sequences, copy numbers and structural variations, somatic mutations, etc. of the ABC transporter superfamily.
4. Materials and Methods
4.1. Patients
The evaluation phase of the study included 105 breast cancer patients, diagnosed in the Institute for the Care for Mother and Child and Medicon, both in Prague and in the Hospital Atlas in Zlin (Czech Republic) over the period of 2006–2013. Of these, 68 patients underwent preoperative (neoadjuvant) treatment with regimens containing 5-fluorouracil, anthracyclines, cyclophosphamide (FAC or FEC), and/or taxanes. The rest received adjuvant postoperative treatment with regimens based on the same drugs. Clinical data of these patients are presented in Table S1.
For the validation phase, we used 802 breast cancer patients, recruited over the period of 2001–2013 from the Institute for the Care for Mother and Child, Medicon, the Motol University Hospital, all in Prague and in the Hospital Atlas in Zlin (all in the Czech Republic). Patients received either neoadjuvant or adjuvant chemotherapy or by hormonal therapy. Clinical data of these patients are presented in Table S3.
More details about the patient recruitment can be found elsewhere [13]. DFS was defined as the time between surgery and first disease relapse including local relapses. Response to NACT was evaluated by the Response Evaluation Criteria in Solid Tumors (RECIST [43]) based on imaging data retrieved from medical records.
Procedures performed in the present study were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Study protocol was approved by the Ethical Commission of the National Institute of Public Health in Prague (approvals no. 9799-4, 15-25618A, and 17-28470A). All patients were informed about the study and those who agreed and signed an informed consent further participated in the study.
4.2. Panel Sequencing—Evaluation Phase
Blood samples were collected during the diagnostic procedures using tubes with K3EDTA anticoagulant and genomic DNA was isolated from human peripheral blood lymphocytes by the standard phenol/chloroform extraction and ethanol precipitation.
In the evaluation phase, raw data for 48 ABC transporter genes and one pseudogene were extracted from the previously published study [13]. Briefly, reads were mapped on reference sequence hg19 using Burrows-Wheeler Alignment (BWA) mem [44], base and indel recalibration and short indels and SNVs discovery was done in the Genome Analysis Toolkit (GATK) [45] and annotation of variants was done using Annovar [46] (for details of the library preparation, target enrichment, data processing, and variant calling, see [13]).
4.3. Genotyping—Validation Phase
In total, 42 genetic variants were selected for the validation phase and assessed using commercially provided competitive allele specific PCR (KASP™) genotyping (LGC Genomics, Hoddesdon, UK) in DNA samples from 802 breast cancer patients. Primers and probes were designed by the service provider. 10% of the samples were analyzed in duplicates for the purpose of the quality control. The genotyping concordance between duplicate samples exceeded 99%.
4.4. Statistical Analyses
In the evaluation phase, DFS was calculated with respect to the groups of patients divided by the genotype (common homozygous, heterozygous, and rare homozygous). The log-rank test for each variant was performed and the Kaplan–Meier plot was generated for visual inspection of gene dosage. We set the study follow-up end to 120 months (10 years) and thus, all subjects with DFS exceeding 120 months were censored. The response of patients to NACT was set to “good” in the case of complete or partial pathological remission (CR/PR) and “poor” for stable or progressive disease (SD/PD). We evaluated associations between genotypes (common homozygous, heterozygous, and rare homozygous) and response using the Pearson chi-square test. Adjusted p-value was calculated for each variant and each of these tests. Adjusted p-value for the log-rank test was based on 100 permutations of original data. A p-value of less than 0.05 after adjustment for multiple testing was considered statistically significant. Variants significantly associating with either DFS or response to NACT in the evaluation phase entered the validation phase of the study.
In the validation phase, the Pearson chi-square test and the log-rank tests were used as described above. For the evaluation of allele effect, recessive, dominant, co-dominant, and additive genetic models were used. Association of variants with transcript levels was assessed by the non-parametric Kruskal–Wallis test. Adjusted p-values were calculated using Benjamini–Hochberg false discovery rate (the FDR test) as a correction for multiple testing [47]. Haplotype analysis was conducted in HaploView 4.2 (Broad Institute, Cambridge, MA, USA). Statistical analyses were conducted using R and the statistical program SPSS v16.0 (SPSS, Chicago, IL, USA).
The sequencing data that support the findings of this study are openly available in Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) under accession no. PRJNA510917.
Acknowledgments
Authors would like to thank Pavel Ostašov from Biomedical Center, Faculty of Medicine in Pilsen, Charles University, Czech Republic for help with computing in R software environment and all participating patients for their kind consent to the study and clinical personnel for outstanding support.
Abbreviations
| ABC | ATP-binding cassette |
| DFS | Disease-free survival |
| FDR | false discovery rate |
| LOF | Loss-of-function |
| MAF | minor allele frequency |
| NGS | next generation sequencing |
| SNP | single nucleotide polymorphism |
| SNV | single nucleotide variant |
| UTR | untranslated regions |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/24/9556/s1.
Author Contributions
Conceptualization, P.S., V.H., and R.V.; methodology, V.H.; software, P.S. and V.H.; validation, P.S., V.B., and V.H.; resources, R.K., K.K., D.V., and J.G.; data curation, R.K., K.K., D.V., and J.G.; writing—original draft preparation, P.S. and V.H.; writing—review and editing, all authors; visualization, P.S., and V.H.; supervision, P.S.; project administration, P.S., V.H., and D.V.; funding acquisition, P.S., V.H., and V.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Czech Medical Council, grant number 17-28470A to P.S., the Czech Ministry of Education, Youth and Sports, grant number CZ.02.1.01/0.0/0.0/16_013/0001634 to V.H., the Grant Agency of Charles University, grant number UNCE/MED/006 to V.B. and the Grant Agency of the Czech Republic, grant number 19-03063S to P.S.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18:452. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szakacs G., Paterson J.K., Ludwig J.A., Booth-Genthe C., Gottesman M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 4.Theodoulou F.L., Kerr I.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015;43:1033–1040. doi: 10.1042/BST20150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasello M., Giudice A.M., Scotlandi K. The ABC subfamily A transporters: Multifaceted players with incipient potentialities in cancer. Semin. Cancer Biol. 2020;60:57. doi: 10.1016/j.semcancer.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Hedditch E.L., Gao B., Russell A.J., Lu Y., Emmanuel C., Beesley J., Johnatty S.E., Chen X., Harnett P., George J., et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J. Natl. Cancer Inst. 2014:106. doi: 10.1093/jnci/dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenna J.G., Taskar K.S., Battista C., Bourdet D.L., Brouwer K.L.R., Brouwer K.R., Dai D., Funk C., Hafey M.J., Lai Y., et al. Can Bile Salt Export Pump Inhibition Testing in Drug Discovery and Development Reduce Liver Injury Risk? An International Transporter Consortium Perspective. Clin. Pharm. 2018;104:916. doi: 10.1002/cpt.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole S.P. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J. Biol. Chem. 2014;289:30880–30888. doi: 10.1074/jbc.R114.609248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slot A.J., Molinski S.V., Cole S.P. Mammalian multidrug-resistance proteins (MRPs) Essays Biochem. 2011;50:179–207. doi: 10.1042/bse0500179. [DOI] [PubMed] [Google Scholar]
- 10.Szakacs G., Abele R. An inventory of lysosomal ABC transporters. FEBS Lett. 2020 doi: 10.1002/1873-3468.13967. [DOI] [PubMed] [Google Scholar]
- 11.Chatuphonprasert W., Jarukamjorn K., Ellinger I. Physiology and Pathophysiology of Steroid Biosynthesis, Transport and Metabolism in the Human Placenta. Front. Pharm. 2018;9:1027. doi: 10.3389/fphar.2018.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerovac M., Tampé R. Control of mRNA Translation by Versatile ATP-Driven Machines. Trends Biochem. Sci. 2019;44:167. doi: 10.1016/j.tibs.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Hlavac V., Kovacova M., Elsnerova K., Brynychova V., Kozevnikovova R., Raus K., Kopeckova K., Mestakova S., Vrana D., Gatek J., et al. Use of Germline Genetic Variability for Prediction of Chemoresistance and Prognosis of Breast Cancer Patients. Cancers. 2018;10:511. doi: 10.3390/cancers10120511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hlavac V., Brynychova V., Vaclavikova R., Ehrlichova M., Vrana D., Pecha V., Kozevnikovova R., Trnkova M., Gatek J., Kopperova D., et al. The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics. 2013;14:515–529. doi: 10.2217/pgs.13.26. [DOI] [PubMed] [Google Scholar]
- 15.Praest P., Luteijn R.D., Brak-Boer I.G.J., Lanfermeijer J., Hoelen H., Ijgosse L., Costa A.I., Gorham R.D., Lebbink R.J., Wiertz E.J.H.J. The influence of TAP1 and TAP2 gene polymorphisms on TAP function and its inhibition by viral immune evasion proteins. Mol. Immunol. 2018;101:55. doi: 10.1016/j.molimm.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 16.Arora H., Wilcox S.M., Johnson L.A., Munro L., Eyford B.A., Pfeifer C.G., Welch I., Jefferies W.A. The ATP-Binding Cassette Gene ABCF1 Functions as an E2 Ubiquitin-Conjugating Enzyme Controlling Macrophage Polarization to Dampen Lethal Septic Shock. Immunity. 2019;50:418. doi: 10.1016/j.immuni.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H., Xia W., Mo X.B., Lin X., Qiu Y.H., Yi N.J., Zhang Y.H., Deng F.Y., Lei S.F. Gene-Based Genome-Wide Association Analysis in European and Asian Populations Identified Novel Genes for Rheumatoid Arthritis. PLoS ONE. 2016;11:e0167212. doi: 10.1371/journal.pone.0167212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fadista J., Oskolkov N., Hansson O., Groop L. LoFtool: A gene intolerance score based on loss-of-function variants in 60 706 individuals. Bioinformatics. 2017;33:471. doi: 10.1093/bioinformatics/btv602. [DOI] [PubMed] [Google Scholar]
- 19.Drean A., Rosenberg S., Lejeune F.X., Goli L., Nadaradjane A.A., Guehennec J., Schmitt C., Verreault M., Bielle F., Mokhtari K., et al. ATP binding cassette (ABC) transporters: Expression and clinical value in glioblastoma. J. Neuro-Oncol. 2018;138:479. doi: 10.1007/s11060-018-2819-3. [DOI] [PubMed] [Google Scholar]
- 20.Hlavata I., Mohelnikova-Duchonova B., Vaclavikova R., Liska V., Pitule P., Novak P., Bruha J., Vycital O., Holubec L., Treska V., et al. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis. 2012;27:187–196. doi: 10.1093/mutage/ger075. [DOI] [PubMed] [Google Scholar]
- 21.Nymoen D.A., Holth A., Hetland Falkenthal T.E., Trope C.G., Davidson B. CIAPIN1 and ABCA13 are markers of poor survival in metastatic ovarian serous carcinoma. Mol. Cancer. 2015;14:44. doi: 10.1186/s12943-015-0317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichler R., Lindner A.K., Comperat E., Obrist P., Schafer G., Todenhofer T., Horninger W., Culig Z., Untergasser G. Amplification of 7p12 Is Associated with Pathologic Nonresponse to Neoadjuvant Chemotherapy in Muscle-Invasive Bladder Cancer. Am. J. Pathol. 2020;190:442. doi: 10.1016/j.ajpath.2019.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Sauna Z.E., Kimchi-Sarfaty C., Ambudkar S.V., Gottesman M.M. Silent polymorphisms speak: How they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 24.Zeng Z., Bromberg Y. Predicting Functional Effects of Synonymous Variants: A Systematic Review and Perspectives. Front. Genet. 2019;10:914. doi: 10.3389/fgene.2019.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S., Shimizu C., Shimoyama T., Takeda M., Ando M., Kohno T., Katsumata N., Kang Y.K., Nishio K., Fujiwara Y. Gene expression profiling of ATP-binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res. Treat. 2006;99:9–17. doi: 10.1007/s10549-006-9175-2. [DOI] [PubMed] [Google Scholar]
- 26.Dvorak P., Pesta M., Soucek P. ABC gene expression profiles have clinical importance and possibly form a new hallmark of cancer. Tumour Biol. 2017;39:1010428317699800. doi: 10.1177/1010428317699800. [DOI] [PubMed] [Google Scholar]
- 27.Xie W., Shui C., Fang X., Peng Y., Qin L. miR-197-3p reduces epithelial-mesenchymal transition by targeting ABCA7 in ovarian cancer cells. 3 Biotech. 2020;10:375. doi: 10.1007/s13205-020-02362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Q., Grimmig T., Gonzalez G., Giobbie-Hurder A., Berg G., Carr N., Wilson B.J., Banerjee P., Ma J., Gold J.S., et al. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J. Biol. Chem. 2018;293:11166. doi: 10.1074/jbc.RA118.003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi J., Naka I., Tsuchiya N. The impact of natural selection on an ABCC11 SNP determining earwax type. Mol. Biol. Evol. 2011;28:849. doi: 10.1093/molbev/msq264. [DOI] [PubMed] [Google Scholar]
- 30.Ishiguro J., Ito H., Tsukamoto M., Iwata H., Nakagawa H., Matsuo K. A functional single nucleotide polymorphism in ABCC11, rs17822931, is associated with the risk of breast cancer in Japanese. Carcinogenesis. 2019;40:537. doi: 10.1093/carcin/bgz005. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa T., Toyoda Y., Yoshiura K., Niikawa N. Pharmacogenetics of human ABC transporter ABCC11: New insights into apocrine gland growth and metabolite secretion. Front. Genet. 2012;3:306. doi: 10.3389/fgene.2012.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo Y., Kotova E., Chen Z.S., Lee K., Hopper-Borge E., Belinsky M.G., Kruh G.D. MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2′,3′-dideoxycytidine and 9′-(2′-phosphonylmethoxyethyl)adenine. J. Biol. Chem. 2003;278:29509–29514. doi: 10.1074/jbc.M304059200. [DOI] [PubMed] [Google Scholar]
- 33.Oba T., Izumi H., Ito K.I. ABCB1 and ABCC11 confer resistance to eribulin in breast cancer cell lines. Oncotarget. 2016;7:70011. doi: 10.18632/oncotarget.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasui K., Mihara S., Zhao C., Okamoto H., Saito-Ohara F., Tomida A., Funato T., Yokomizo A., Naito S., Imoto I., et al. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res. 2004;64:1403–1410. doi: 10.1158/0008-5472.CAN-3263-2. [DOI] [PubMed] [Google Scholar]
- 35.Low F.G., Shabir K., Brown J.E., Bill R.M., Rothnie A.J. Roles of ABCC1 and ABCC4 in Proliferation and Migration of Breast Cancer Cell Lines. Int. J. Mol. Sci. 2020;21:7664. doi: 10.3390/ijms21207664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xavier B.M., Zein A.A., Venes A., Wang J., Lee J.-Y. Transmembrane Polar Relay Drives the Allosteric Regulation for ABCG5/G8 Sterol Transporter. Int. J. Mol. Sci. 2020;21:8747. doi: 10.3390/ijms21228747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biedler J.L., Riehm H. Cellular resistance to actinomycin D in Chinese hamster cells in vitro: Cross-resistance, radioautographic, and cytogenetic studies. Cancer Res. 1970;30:1174–1184. [PubMed] [Google Scholar]
- 38.Madrid-Paredes A., Cañadas-Garre M., Sánchez-Pozo A., Expósito-Ruiz M., Calleja-Hernández M.Á. ABCB1 gene polymorphisms and response to chemotherapy in breast cancer patients: A meta-analysis. Surg. Oncol. 2017;26:473. doi: 10.1016/j.suronc.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Yamada A., Nagahashi M., Aoyagi T., Huang W.C., Lima S., Hait N.C., Maiti A., Kida K., Terracina K.P., Miyazaki H., et al. ABCC1-Exported Sphingosine-1-phosphate, Produced by Sphingosine Kinase 1, Shortens Survival of Mice and Patients with Breast Cancer. Mol. Cancer Res. 2018;16:1059. doi: 10.1158/1541-7786.MCR-17-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeil A.M., Vulsteke C., Paridaens R., Dieudonné A.S., Pettengell R., Hatse S., Neven P., Lambrechts D., Szucs T.D., Schwenkglenks M., et al. Multivariable regression analysis of febrile neutropenia occurrence in early breast cancer patients receiving chemotherapy assessing patient-related, chemotherapy-related and genetic risk factors. BMC Cancer. 2014;14:201. doi: 10.1186/1471-2407-14-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Q., Zhou Y., Winter S., Büttner F., Schaeffeler E., Schwab M., Lauschke V.M. Germline variant burden in multidrug resistance transporters is a therapy-specific predictor of survival in breast cancer patients. Int. J. Cancer. 2020;146:2475. doi: 10.1002/ijc.32898. [DOI] [PubMed] [Google Scholar]
- 42.Reustle A., Fisel P., Renner O., Büttner F., Winter S., Rausch S., Kruck S., Nies A.T., Hennenlotter J., Scharpf M., et al. Characterization of the breast cancer resistance protein (BCRP/ABCG2) in clear cell renal cell carcinoma. Int. J. Cancer. 2018;143:3181. doi: 10.1002/ijc.31741. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz L.H., Litiere S., de Vries E., Ford R., Gwyther S., Mandrekar S., Shankar L., Bogaerts J., Chen A., Dancey J., et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur. J. Cancer. 2016;62:132. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y., Hochberg Y. Controlling the false discovery rate A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.