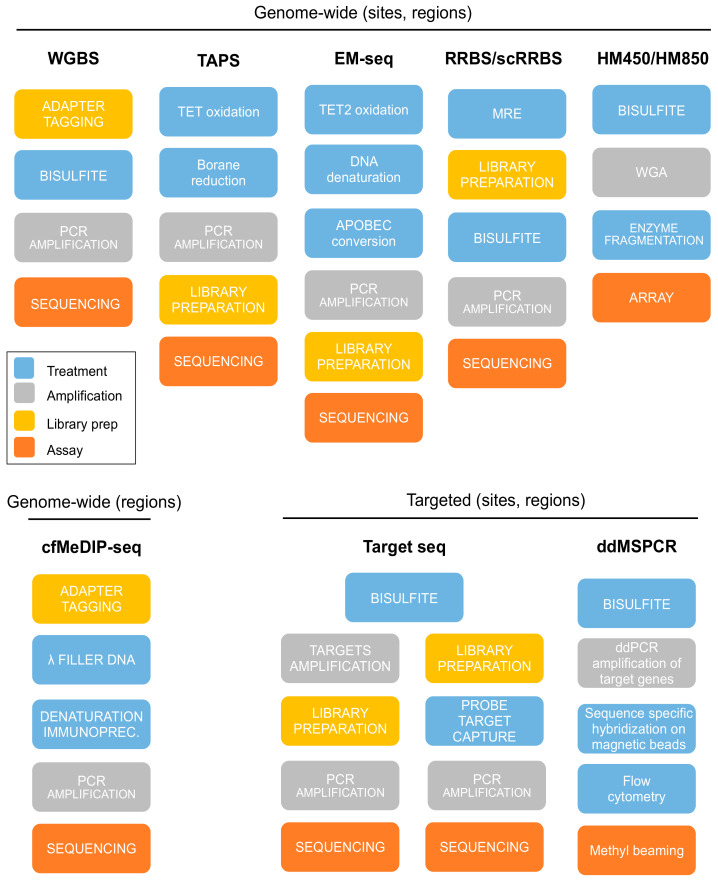
Figure 2.
Summary of the experimental assays for DNA-methylation evaluation. Genome-wide techniques include methods able to perform site and region wise analysis, such as whole genome bisulfite sequencing (WGBS), TET-assisted pyridine borane sequencing (TAPS), enzymatic methyl-sequencing (EM-seq), reduced representation of bisulfite sequencing (RRBS, single-cell RRBS (scRRBS)), and infinium methylation arrays (HM450, HM850), and methods analyzing DNAm regions only, such as circulating free methylated DNA immunoprecipitation sequencing (cfMeDIP–seq). Targeted methods include target bisulfite sequencing (target bisulfite seq) and PCR-based assays, including methylation specific PCR (MSPCR) and droplet digital methylation specific PCR (ddMSPCR). MRE: methylation restriction enzyme, WGA: whole genome amplification.