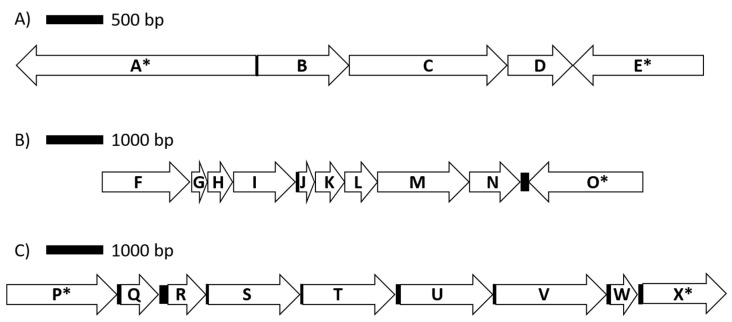
Figure 3.
Genomic organization for genes encoding proteins involved in lactate utilization (A), the ATP synthase complex (B) and arabinose metabolism (C). Coding sequences are represented by arrows, while intergenic regions are shown as solid boxes. Available contig sequences permitting, genes flanking coding sequences of interest are indicated by (*). Annotated protein/enzymes displayed are (A) DNA topoisomerase; (B) L-lactate dehydrogenase, Fe-S oxidoreductase; (C) lactate utilization domain protein; (D) lactate utilization domain protein; (E) DegT/DnrJ/EryC1/StrS aminotransferase family; (F) F0F1 ATP synthase subunit beta; (G) F0F1 ATP synthase subunit epsilon; (H) hypothetical protein; (I) F0F1 ATP synthase subunit A; (J) F0F1 ATP synthase subunit C; (K) F0F1 ATP synthase subunit B; (L) F0F1 ATP synthase subunit delta; (M) F0F1 ATP synthase subunit alpha; (N) F0F1 ATP synthase subunit gamma; (O) fructose 1,6 bisphosphatase; (P) sodium solute symporter; (Q) nudix hydrolase; (R) L-ribulose-5-phosphate 4-epimerase; (S) L-arabinose isomerase; (T) FGGY-family carbohydrate kinase; (U) alpha-N-arabinofuranosidase; (V) transketolase; (W) ribose 5 phosphate isomerase; (X) pyruvate kinase.