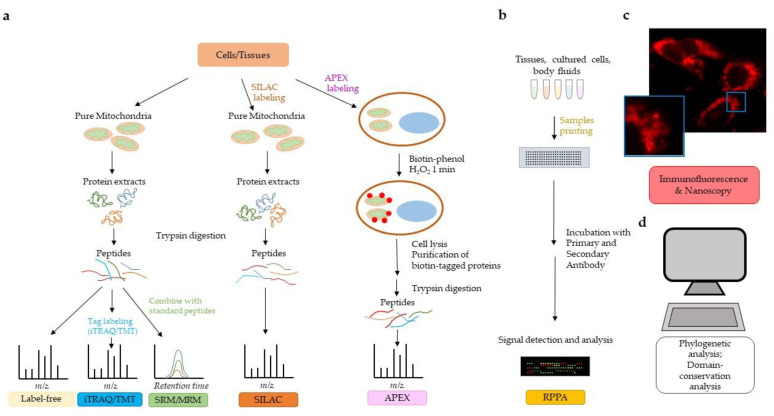
Figure 1.
Strategies to investigate the mitochondrial proteome. (a) Mass spectrometry (MS) technology allows qualitative and quantitative analysis of the mitochondrial proteome. Different labeling methods can be used to quantify the mitochondrial proteome (see text for details). Label-free and iTRAQ/TMT allow discovery of proteins in a biological sample. SRM/MRM and SILAC are MS technologies that allow proteome quantification. APEX bypasses the need for organelle purification. (b) Reverse phase protein array (RPPA) allows for the quantification of proteins from relatively small amounts of biological samples (tissues, cell cultures, body fluids) and requires the availability of highly specific primary antibodies. (c) Immunofluorescent microscopy/nanoscopy is widely used to assess and/or confirm mitochondrial localization of proteins. The immunofluorescent image shows human pancreatic cancer cells stained with a primary antibody to pyruvate dehydrogenase followed by Alexa Fluor 568 secondary antibody. Nanoscopy can provide a higher resolution on subcellular compartmentalization than traditional immunofluorescence, as detailed in the text. (d) Bioinformatic analyses of mitochondrial targeting sequences, phylogenetic analyses, domain conservation analyses, and homology comparisons to previously identified mitochondrial proteins may predict localization of proteins in mitochondria.