Abstract
Drought is one of the major environmental stresses adversely affecting crop productivity worldwide. Precise characterization of genes involved in drought response is necessary to develop new crop varieties with enhanced drought tolerance. Previously, we identified 66 drought-induced miRNAs in rice plants. For the further functional investigation of the miRNAs, we applied recombinant codon-optimized Cas9 (rCas9) for rice with single-guide RNAs specifically targeting mature miRNA sequences or sites required for the biogenesis of mature miRNA. A total of 458 T0 transgenic plants were analyzed to determine the frequency and type of mutations induced by CRISPR/rCas9 on 13 independent target miRNAs. The average mutation frequency for 13 genes targeted by single guide RNAs (sgRNAs) in T0 generation was 59.4%, including mono-allelic (8.54%), bi-allelic (11.1%), and hetero-allelic combination (39.7%) mutations. The mutation frequency showed a positive correlation with Tm temperature of sgRNAs. For base insertion, one base insertion (99%) was predominantly detected in transgenic plants. Similarly, one base deletion accounted for the highest percentage, but there was also a significant percentage of cases in which more than one base was deleted. The deletion of more than two bases in OsmiR171f and OsmiR818b significantly reduced the level of corresponding mature miRNAs. Further functional analysis using CRISPR/Cas9-mediated mutagenesis confirmed that OsmiR818b is involved in drought response in rice plants. Overall, this study suggests that the CRISPR/rCas9 system is a powerful tool for loss-of-function analysis of miRNA in rice.
Keywords: CRISPR/rCas9, drought-responsive miRNAs, INDELs, Oryza sativa, rice protoplast, sgRNAs
1. Introduction
Targeted genome editing with site-directed nucleases (SDNs) is a promising tool for both basic and applied biological research. The SDNs, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly-interspaced short palindromic repeats (CRISPR)-associated 9 (Cas9) system, induce targeted DNA double-strand breaks (DSBs) and subsequently trigger DNA repair through non-homologous end-joining (NHEJ) or homologous recombination (HR) pathway [1,2,3]. SDN-mediated genome editing facilitates the development of transgene-free new crop varieties because the changes of the genome sequence are able to be separated from the transgene.
Among three representative SDNs, CRISPR/Cas9 is regarded as an efficient system to introduce mutations and gene fragments into the plant genome [3,4]. The CRISPR/Cas9 system is composed of a Cas9 endonuclease, a CRISPR RNA (crRNA), and a trans-acting crRNA (tracrRNA) isolated from Streptococcus pyogenes [1]. A crRNA is responsible for targeting a specific DNA site, and a tracrRNA provides a scaffold that is recognized by a Cas9 protein. The crRNA is base-paired to tracrRNA and form a two-RNA structure that directs the Cas9 to the target site [5]. A chimeric single-guide RNA (sgRNA), which is a fusion of a CRISPR RNA (crRNA) and a trans-acting crRNA (tracrRNA), also successfully directs Cas9 for sequence-specific DNA cleavage [6]. The CRISPR/Cas9 system recognizes 20 specific nucleotides followed by a protospacer-adjacent motif (PAM) sequence and cleaves DNA at approximately three base pairs (bps) upstream of the PAM sequence. During the repair processes, variable sizes of base insertions and deletions (indels) occur at target sites, resulting in site-specific changes of genetic information. Thus, the proper design of sgRNA is sufficient to induce mutations on the specific location of the genome. In addition, the CRISPR/Cas9 system can simultaneously edit multiple target sites by using multiple gRNAs encoded in a single CRISPR array [7]. Based on these advantages, the CRISPR/Cas9 system is extensively used for the functional characterization of genes, especially in plants [4,7,8].
microRNA (miRNA) is a class of small non-coding RNAs that directly regulate the functions of specific messenger RNAs through transcriptional or translational repression [9,10,11]. miRNA is transcribed into a form of primary miRNA and processed into 20–24 nt small mature miRNAs through the actions of multiple components such as Dicer-like RNase III endonucleases (DCLs) [12]. Gain- and loss-of-functional approaches are the most effective strategies to characterize the function of a particular gene [13]. Given the nature of miRNA that is transcribed from the genome, precise determination of a transcript unit is required for gain-of-functional approaches by overexpressing it. Artificial RNA can be also directly expressed under the control of appropriate promoters [13,14]. On the other hand, loss-of-function analysis of miRNA is difficult due to the lack of appropriate knockout mutants. Loss-of-function mutation of miRNA is difficult to achieve by insertional mutagenesis due to the small sequence size and insufficient coverage of inserts in rice. For this reason, post-transcriptional approaches that sequester or degrade miRNAs have been used for the loss-of-function analysis of miRNA [15,16,17], but these approaches often produced considerable variations in the degree of inhibition on miRNA function [18]. Recently, CRISPR/Cas9 has been spotlighted as an alternative option to overcome these limitations [4,19]. Here, we aimed to generate a loss-of-function mutant of 12 independent miRNAs related to drought responses in rice using miRNA-specific sgRNA [20] and recombinant codon-optimized Cas9 (rCas9) for rice. The large-scale analysis of transgenic plants expressing the CRISPR/rCas9 system provides technical and biological information that is helpful for reversing genetic approaches for the functional analysis of miRNA using the CRISPR/rCas9 system in rice.
2. Results
2.1. Functional Validation of Rice Codon-Optimized rCas9 Using Rice Protoplasts
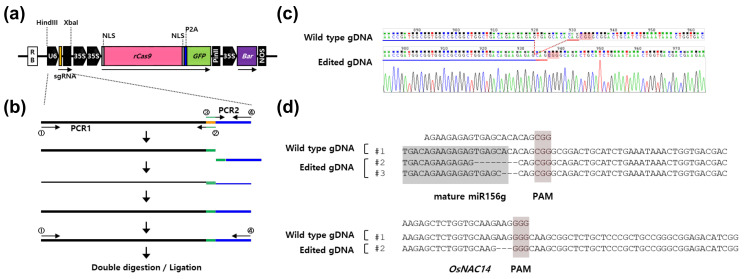
To accommodate the CRISPR/rCas9 system in rice, we constructed a binary vector expressing single guide RNA (sgRNA) under the control of rice U6 promoter and recombinant codon-optimized Streptococcus pyogenes Cas9 (rCas9) under the control of CaMV 35S promoter (pSB11_U6::sgRNA-35S::rCas9) (Figure 1a). The target-specific gRNA sequence was inserted between the rice U6 promoter and the sgRNA scaffold by overlapping PCR (Figure 1b). To test whether the recombinant CRISPR/rCas9 system worked properly and could be used to specifically modify an endogenous genome in rice, we designed two sgRNAs specifically targeting the OsmiR156g and OsNAC14 locus, respectively. The final constructs were transiently expressed in rice leaf protoplasts. To identify mutations induced by transient expression of the CRISPR/Cas9 system, sgRNAs targeting genomic regions were analyzed by cloning PCR products into the TA cloning vector followed by sequencing (Figure 1c). Sequencing analysis revealed that de novo mutations were generated on the OsmiR156 gene targeted by sgRNA, and two different deletion mutations were identified in which three or eight bases were removed out of 10 sequencing results (Figure 1d). Similarly, three base pair deletions were found from the OsNAC14 locus targeted by sgRNA (Figure 1d). The deletions were located in close proximity to the predicted cleavage site of the Cas9/sgRNA complex (three to four bps downstream of the PAM sequence or immediately adjacent to the PAM sequence), indicating that the deletions of target sequences could be caused by rCas9-directed DNA cleavage and followed the non-homologous end-joining repair pathway.
Figure 1.
Construction of binary vectors and confirmation of clustered regularly-interspaced short palindromic repeats (CRISPR)/ recombinant codon-optimized Cas9 (rCas9) system in rice protoplasts. (a) Diagram of recombinant CRISPR/rCas9 constructs. A rice codon-optimized Streptococcus pyogenes Cas9 (rCas9) fusing with N-terminal and C-terminal nuclear localization signal (NLS), self-cleaving 2A peptide (P2A), and GFP were inserted downstream of dual Cauliflower mosaic virus 35S (CaMV 35S) promoters. The rice U6 promoter and single-guide RNA (sgRNA) were cloned between the right border (RB) and CaMV 35S promoters. Bar, phosphinothricin N-acetyltransferase; PinII, potato proteinase inhibitor II terminator; NOS, nopaline synthase terminator; LB, left border driven by 35S promoter. (b) Construction of the pU6:sgRNA cassette using overlapping PCR. To introduce a specific sequence between the U6 promoter and guide RNA scaffold sequence, a specific sequence was incorporated at the 5′ ends of guide RNA scaffold specific forward primer (③). The reverse complementary sequence of the specific sequence was added at the 5′ end of the U6 promoter-specific reverse primer (②). Two PCR products (PCR1 and PCR2) were annealed together through base pairing between the inserted specific sequences, and each strand was extended by a sequence that was complementary to the sequence it was to be joined to, producing a dsDNA template. An additional round of PCR was carried out to amplify the pU6:sgRNA cassette. The final PCR product was inserted into a CRISPR/rCas9 vector through HindIII and XbaI restriction sites. (c) The rice protoplasts were transfected with a CRISPR/rCas9 vector containing sgRNA specific for OsmiR156g. A representative chromatogram was obtained from direct PCR sequencing analysis of the transfected protoplasts. The mutated target region is indicated with red dotted line. The protospacer adjacent motif (PAM) site is underlined in red. (d) The rice protoplasts were transfected with a CRISPR/rCas9 vector containing sgRNA specific for OsmiR156g or OsNAC14. Genomic DNA extracted from the protoplasts was used for amplification of the genomic region targeted by sgRNA by PCR reaction. The PCR products were then subcloned into the TA cloning vector for sequencing analysis. The mature miR156g sequence and PAM site are highlighted with gray and red boxes, respectively.
2.2. Genome Editing Patterns Generated by rCas9
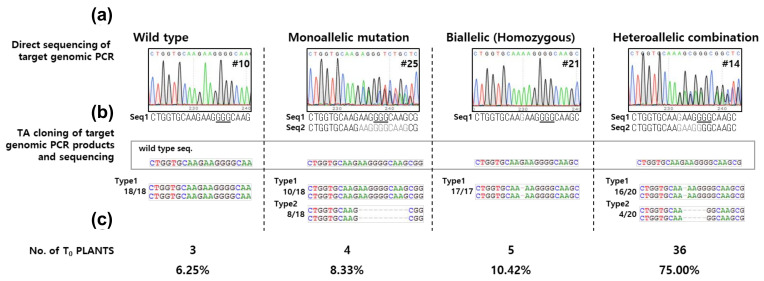
To confirm that stable expression of rCas9 also induces mutation in rice, we generated transgenic rice plants expressing rCas9 under the control of 35S promoter and OsNAC14-specific sgRNA under the control of U6 promoter through Agrobacterium-mediated transformation (CRISPR/rCas9OsNAC14). Mutation patterns in the sgRNA targeting site were confirmed by targeted sequencing analysis of PCR products (Figure 2a and Figure S1). The accuracy of the targeted sequencing method was further confirmed by TA-cloning-coupled sequencing of the sgRNA targeting region (Figure 2b). The sequencing analysis of the 48 T0 transgenic plants revealed that the CRISPR/rCas9 system successfully induced mutations on the sgRNA targeting site. Among the analyzed 48 T0 plants, 45 transgenic plants contained indels at the target sites, four transgenic plants contained monoallelic mutations, and 41 transgenic plants contained biallelic or heteroallelic combination mutations. Various types of mutations were found in the analyzed CRISPR/rCas9OsNAC14 plants. Two bases were frequently deleted in the analyzed CRISPR/rCas9OsNAC14 plants (Figure S2). In the case of base insertion, one base insertion was predominantly detected from the 48 plants. These results indicate that CRISPR/rCas9 can be used for site-specific mutagenesis in rice.
Figure 2.
Confirmation of the CRISPR/rCas9 system in rice. Genomic DNA was extracted from the 48 T0 transgenic plants expressing rCas9 and sgRNA specific for OsNAC14 and used for sequencing analysis. (a) Representative chromatograms obtained from the sequencing analysis of PCR products. (b) Type of mutations on the sgRNA target site determined through TA-cloning-coupled sequencing analysis. (c) The number of T0 plants showing an indicated type of mutation. Wild type, non-mutated transgenic plants; monoallelic mutation, only one allele is mutated; biallelic, the two alleles are mutated; homozygous, the two alleles have the same mutations; heterozygous, the two alleles have different mutations.
2.3. The Efficiency of CRISPR/rCas9-Mediated Mutagenesis on miRNA Genes
To precisely evaluate the efficiency of rCas9-driven mutation, we chose 12 rice drought-responsive miRNAs as targets for CRISPR/rCas9-mediated mutagenesis. A total of 458 T0 transgenic rice plants was used for analyzing mutation frequency and indel patterns on the sgRNA-targeting sites. Sequencing analysis showed that 59.4% (272 out of 458) of transgenic plants contained mutations on their target sites (Table 1). The highest mutation efficiency was 100% for miR399K and the lowest mutation efficiency was 3.0% for miR816. The majority of mutation types were biallelic mutations (50.8%), and 8.5% of the transgenic plants contained monoallelic mutations. Among biallelic mutations, homozygous and heterogeneous mutations accounted for 21.9% and 78.1%, respectively.
Table 1.
List of target genes and mutation patterns in corresponding transgenic plants.
| Target Gene | gRNA | GC(%) | Tm | WT | Mono-Allelic | Biallelic | Total Plants | Mutation Rate (%) | |
|---|---|---|---|---|---|---|---|---|---|
| Homo | Hetero | ||||||||
| miR399d | TCACCAAAACGGCCTGCCAAAGG | 55 | 73.0 | 40 | 6 | 46 | 13.0% | ||
| miR418 | AATTCCACCGTGGTCCCTGGAGG | 60 | 70.5 | 26 | 6 | 10 | 42 | 38.1% | |
| miR156d | AGAGTGAGCACACGGCGTGATGG | 60 | 70.1 | 3 | 8 | 37 | 48 | 93.8% | |
| miR399e | TGCCCAGCAATGCAACTTTGCGG | 50 | 69.6 | 14 | 14 | 1 | 3 | 32 | 56.3% |
| miR399i | TGCTAGCCTTTCCCTGCCAAAGG | 55 | 69.2 | 3 | 6 | 8 | 4 | 21 | 85.7% |
| miR169f | AAGAGCTGATTCGGTAGCCAAGG | 50 | 64.2 | 7 | 3 | 10 | 29 | 49 | 85.7% |
| miR171f | TTGGCATGGTTCAATCAAACCGG | 40 | 63.7 | 21 | 1 | 3 | 24 | 49 | 57.1% |
| OsNAC14 | AAGAGCTCTGGTGCAAGAAGGGG | 50 | 61.8 | 3 | 4 | 5 | 36 | 48 | 93.8% |
| miR156g | GAAGAGAGTGAGCACACAGCGGG | 55 | 61.0 | 1 | 2 | 3 | 12 | 18 | 94.4% |
| miR399k | GGTTACCAGACTACTGCCAAAGG | 50 | 59.7 | 3 | 7 | 20 | 30 | 100.0% | |
| miR818b | ATCCAAAATCCCTTATATTATGG | 25 | 52.5 | 17 | 2 | 19 | 10.5% | ||
| miR814a | ACTTCATAGTACAACGAATCTGG | 35 | 51.3 | 19 | 3 | 1 | 23 | 17.4% | |
| miR816 | ATATTTTACTACAACGAATCTGG | 25 | 48.2 | 32 | 1 | 33 | 3.0% | ||
| Total | 186 | 39 | 51 | 182 | 458 | ||||
| 40.6% | 8.5% | 11.1% | 39.7% | ||||||
Mutation data were obtained from sequencing analysis of a total of 458 T0 transgenic plants transformed with CRISPR/rCas9. sgRNA, target sequence (black) and PAM sequence (red); GC (%), GC content of 20 bp sgRNA sequence; Tm, melting temperature of 20 bp sgRNA sequence; WT, non-mutated transgenic plants; mono-allelic, only one allele is mutated; bi-allelic, the two alleles are mutated; homo, the two alleles have the same mutations; hetero, the two alleles have different mutations; total plants, number of transgenic plants used for the analysis; mutation rate (%), the ratio of transgenic plants with mutations from total transgenic plants.
2.4. Patterns and Position of CRISPR/rCas9-Mediated Mutations in T0 Transgenic Rice
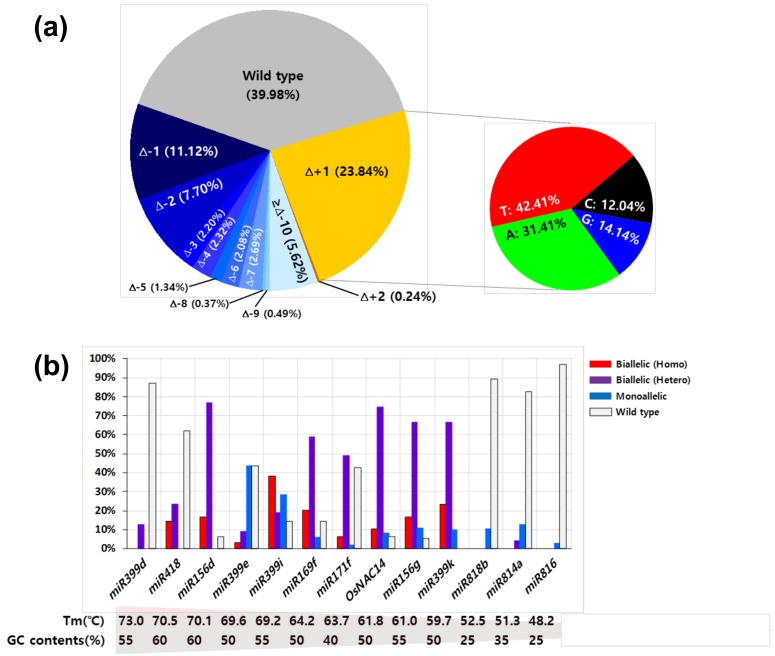
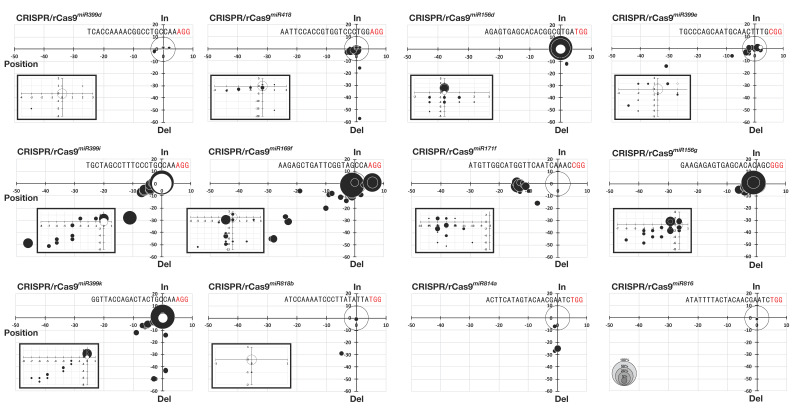
We then analyzed mutation patterns in the transgenic plants. The base deletions and insertions accounted for 35.9% and 24.1%, respectively (Figure 3a). The proportion of one and two base deletions was 52.4% of total base deletions, and more than three base deletions accounted for 47.6%. In the case of base insertion, 99% of target sites with insertion contained single base insertion (Figure 3a). All four bases were inserted into the plant genome, of which thymine (T) or adenine (A) had a high proportion (73.82%) (Figure 3a). We found a potential correlation between Tm temperature and mutation frequency (Figure 3b). Relatively-higher mutation frequency was observed when using sgRNAs with a high Tm temperature than those with low Tm temperature except for the miR399d locus. In addition, frequent biallelic mutations were observed in plants transformed with sgRNAs with high Tm temperatures (Figure 3b). On the other hand, monoallelic mutations were predominantly observed in transgenic plants expressing sgRNAs with Tm temperatures lower than 40% (Figure 3b). Next, we analyzed the correlation between the size of the indel and the position of the mutation (Figure 4). Differently from mutations observed in protoplasts, mutations occurred on not only three bases upstream of the PAM site but also other positions in the transgenic plants. Moreover, several plants carried mutations on the PAM site or even downstream of the PAM site (Figure 4). Large deletions of more than 10 bases were observed in CRISPR/rCas9miR418, CRISPR/rCas9miR156d, CRISPR/rCas9miR399e, CRISPR/rCas9miR399i, CRISPR/rCas9miR169f, CRISPR/rCas9miR171f, CRISPR/rCas9miR399k, and CRISPR/rCas9miR814a transgenic plants. The longest deletion detected was 68 base deletions in CRISPR/rCas9miR418 transgenic plants (Figure 4). These results indicate that the CRISPR/rCas9 system could induce various types of deletions that would be beneficial for the effective elimination of miRNA functions.
Figure 3.
Mutation efficiency and type in T0 transgenic rice plants expressing CRISPR/Cas9. Sequence information of the sgRNA targeting site was collected from 458 T0 transgenic rice plants. (a) The pie chart displays the frequency of each mutation type found in 458 T0 transgenic rice plants. (b) The bar chart indicates the proportion of mutation types in transgenic plants. Tm, melting temperature of 20 bp sgRNA sequence; GC content, GC content of 20 bp sgRNA sequence; bi-allelic (homo), the two alleles have the same mutated (red bar); bi-allelic (hetero), the two alleles have different mutations (purple bar); mono-allelic, only one allele is mutated (blue bar); wild type, plants that had not been mutated (White bar).
Figure 4.
Patterns and positions of CRISPR/rCas9-mediated base insertion and deletion (Iindel) mutations. The graph shows the correlation between the degree of indel (y-axis) and the position where indel occurs (x-axis). The square in the graph is an enlarged image of the area where spots are concentrated. The white circle at the origin indicates the frequency of non-mutated plants.
2.5. Inheritance of rCas9-Mediated Mutation
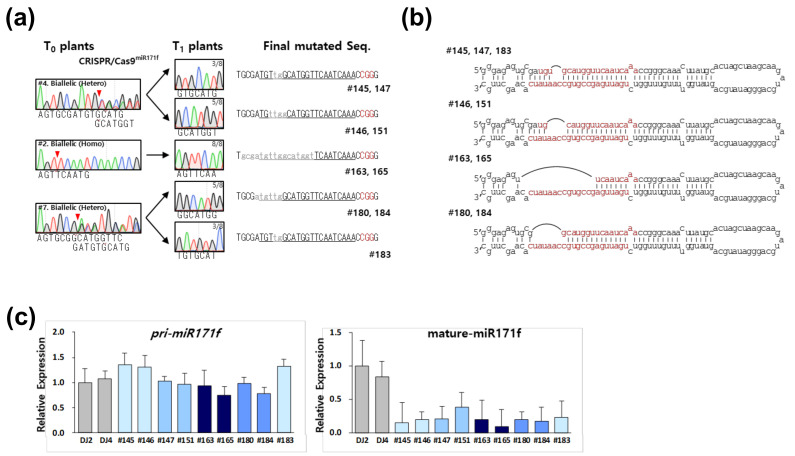
To confirm the stable transmission of rCas9-mediated mutation in T0 plants into its T1 siblings, we chose transgenic plants carrying three different forms of mutations on miR171f. CRISPR/rCas9miR171f #4 transgenic plants contained biallelic mutations composed of deletion of two or four bases in the T0 stage. In the T1 stage, three plants were homozygous mutants with two base deletions while five plants turned out to be homozygous mutants with four base deletions (Figure 5a). Similarly, biallelic mutations in the T0 stage of CRISPR/rCas9miR171f #7 transgenic plants were segregated in the T1 stage. In addition, CRISPR/rCas9miR171f #2 transgenic plants carrying a homozygous mutation in the T0 stage also had a homozygous mutation in the T1 stage (Figure 5a). These results indicate that mutations observed in transgenic plants in the T0 stage are successively inherited into the next siblings.
Figure 5.
Characterization of CRISPR/Cas9miR171f T1 plants with homozygous mutations. (a) Mutation patterns were determined from CRISPR/Cas9miR171f T1 plants. T1 generation obtained from three independent CRISPR/Cas9miR171f transgenic plants was used to investigate the inheritance and segregation of mutations observed in the T0 stage. (b) Prediction of the stem–loop structure of the mutated miR171f. The mature miR171f is shown in red. (c) Expression patterns of pri-miR171f and mature miR171f-5p in non-transgenic and CRISPR/Cas9miR171f transgenic plants.
Differently from a gene that codes protein, miRNA functions as an RNA, thus insertion or deletion of bases is insufficient to guarantee functional impairment of a miRNA. To evaluate the effect of rCas9-driven mutations on the function of miRNAs, we first illustrated the position of mutations on the pre-miR171f sequence (Figure 5b). All four mutations were located on the mature miR171f sequence. The mutations did not affect transcription of pre-miR171f transcripts as evidenced by qPCR analysis (Figure 5c). However, the mutations significantly reduced the level of mature OsmiR171f-5p (Figure 5c). These results suggest that deletions of mature miR171 sequence affect its transcript level in rice.
2.6. Function of the Drought-Induced OsmiR818b on Drought Responses in Rice Plants
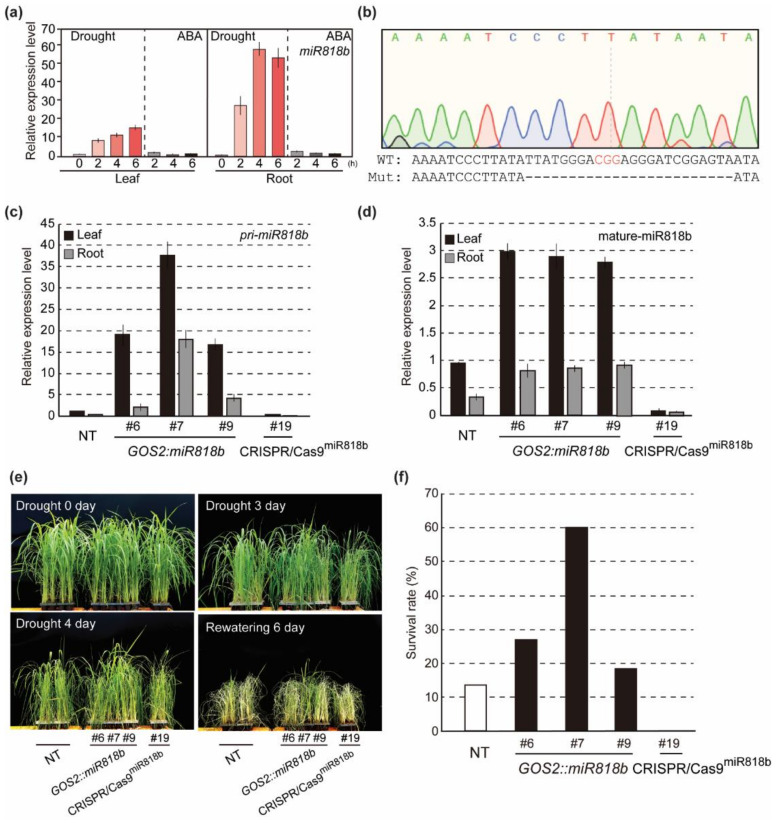
To confirm that the CRISPR/Cas9-mediated mutagenesis can be used for functional analysis of miRNAs, we used the drought-responsive OsmiR818b for further analysis. We first investigated expression patterns of OsmiR818b in response to drought. OsmiR818b expression was induced by drought treatments in both leaves and roots of rice plants (Figure 6a). On the other hand, OsmiR818b expression was rarely changed by abscisic acid (ABA) treatments (Figure 6a). These data indicate that OsmiR818b expression is induced by drought in an ABA-independent pathway. To address function of OsmiR818b, we generated OsmiR818b-overexpressing transgenic plants (GOS2::miR818b) and CRIPSR/Cas9miR818b mutant plants. The CRIPSR/Cas9miR818b #19 mutant plants contained a biallelic mutation composed of a 24 base deletion (Figure 6b). Expression analysis revealed that both primary (pri-) and mature-OsmiR818b were highly expressed in OsmiR818b-overexpressing plants while levels of pri- and mature-OsmiR818b was reduced in CRIPSR/Cas9miR818b mutant plants (Figure 6c,d). To investigate effect of the altered expression of OsmiR818b on drought response in rice plants, we treated drought stress on OsmiR818b-overexpressing and CRIPSR/Cas9miR818b mutant plants as well as non-transgenic (NT) control plants, and monitored drought-induced visual symptoms. Drought induced symptom such as leaf rolling and wilting appeared earlier in CRIPSR/Cas9miR818b mutant plants compared to those in NT control plants (Figure 6e). in addition, CRIPSR/Cas9miR818b mutant plants showed lower recovery rate compared to NT plants after being relieved from drought stress through re-watering (Figure 6f). On the other hand, OsmiR818b-overexpressing plants were relatively more tolerant to drought treatment than NT plants (Figure 6e,f). These result suggest that OsmiR818b participates in drought response of rice plants and increase of OsmiR818b expression improves drought tolerance in rice plants.
Figure 6.
Drought response of OsmiR818b-overexpressing and CRISPR/Cas9miR818b plants. (a) Expression patterns of pri-miR818b in response to drought and abscisic acid (ABA) treatments. Two-week-old non-transgenic rice (O. sativa cv. Dongjin) seedlings were treated with drought and 100 μM ABA. The plants were harvested at the indicated time points after treatments. (b) Mutation pattern on OsmiR818b sequence in CRISPR/Cas9miR818b #19 transgenic plants. (c) Expression level of pri-OsmiR818b and (d) mature-OsmiR818b in OsmiR818b-overexpressing (GOS2:miR818b) and CRISPR/Cas9miR818b #19 transgenic plants. (e) The responses of the transgenic plants during drought treatments. One-month-old non-transgenic (NT), OsmiR818b-overexpressing (GOS2:miR818b) and CRISPR/Cas9miR818b #19 mutant plants were exposed to drought stress for 4 days, followed by re-watering for 6 days. (f) The survival rate of the transgenic plants 6 days after re-watering (n = 30).
3. Discussion
Genome editing efficiency in plants is largely determined by several crucial factors including the expression level of Cas9 and sgRNA [21,22]. Codon optimization is one of the feasible options to increase the translation of recombinant protein in plants. For this reason, several plant codon-optimized Cas9 have been introduced into various plants [23,24]. Here, we tested mutation efficiency using recombinant rice codon-optimized Cas9 (rCas9). Our data suggest that the CRISPR/rCas9 system can induce mutations on target sites with high efficiency in rice (Table 1). rCas9 successfully induced mutations on plant genomes targeted by sgRNA in both transient and stable expression systems (Figure 1 and Figure 2). Sequence analysis of 458 transgenic plants at the T0 stage revealed that 59.4% of the tested plants carried monoallelic or biallelic mutations (Table 1). Biallelic mutations were found from 66.9% of the transgenic plants with mutations. In addition, mutation efficiency was greater than 85.0% in 6 out of the 13 tested miRNAs (Table 1). Moreover, the probability of occurrence of homozygous mutations was 11.1% (Table 1). It has been reported that the introduction of SpCas9 with sgRNA showed a mutation induction rate of 44.4% in rice, and 3.8% of transgenic plants carrying homozygous mutations [25]. Thus, the CRISPR/rCas9 system used in this study can be a tool for efficient targeted mutagenesis in rice.
The mutation frequency was related to the Tm temperature of the target sequences. Target sequences with Tm temperature over 53 °C except for miR399d showed significantly-higher mutation frequency than those with low Tm temperature (Figure 3b). A similar correlation between mutation frequency and GC ratio has been reported in several organisms [8,26,27]. It has also been reported that the existence of purine residues at the end of the sgRNA is important for genome editing efficiency [8]. Our analysis showed that the contribution of purine residues in sgRNA on genome editing efficiency seems relatively minor compared to that of Tm temperature.
CRISPR/rCas9-mediated mutations are generally detected three bases upstream of the PAM site because Cas9 binds to sgRNA and cleaves three bases upstream of the PAM site [8]. Consistently, our data showed that all detected mutations generated by transient expression of CRISPR/rCas9 in protoplasts were located three bases upstream of the PAM sequence (Figure 1c,d). However, stable expression of CRISPR/rCas9, even with the same constructs, caused mutations in a wider range of regions including three bases upstream of the PAM site (Figure 2 and Figure 4). It is not clear how these differences were generated between two different expression systems. One possible explanation is that NHEJ activity or stability of truncated double-strand DNA might be different between protoplasts and calluses. Similar to our data, Shan et al. reported that the TALEN system produced longer deletions by stable expression in calluses compared with transient expression in protoplasts of rice and Brachypodium [28]. Further investigations will be helpful to understand the difference in mutation types that occurred by transient and stable expression of molecular scissors.
Strategies for miRNA knockout are essential for studying miRNA function in plants [29]. The major loss-of-function technologies in miRNA research include miRNA-specific antisense inhibitors, miRNA sponges, and genetic knockouts [30]. Among them, a genetic knockout is the most reliable technique to determine the functions of miRNA. Hitherto, the CRISPR/Cas9 system has been exclusively applied in human, mouse, or zebrafish cell lines to knockout miRNA genes [31,32,33,34]. Similarly, various attempts have been reported to create miRNA knockout plants using the CRISPR/Cas9 system for the functional characterization of miRNA in plants [19,29]. Mutations on mature miRNA sequences or flanking regions which are important for miRNA processing are crucial for successful knockout of miRNA. In addition, the deletion of multiple bases will increase the chance of miRNA knockout production. Our data indicate that stable expression of the CRISPR/rCas9 system in rice generated large deletions on miRNA sequences targeted by sgRNAs (Figure 4). For example, the CRISPR/rCas9 system induced 2 to 16 base deletions on the miR171f gene (Figure 5a). The deletions changed the level of mature miRNA171f but not the transcription level of pri-miR171f (Figure 5c). Moreover, deletion of 24 bases on the miR818b significantly reduced levels of both pri- and mature-miR818b (Figure 6c,d). The functional analysis using CRISPR/Cas9mir818b mutant and miR818b-overexpressing plants showed that OsmiR818b is involved in drought tolerance in rice plants (Figure 6e,f). These data indicate that a deletion within the pre-miRNA regions by CRISPR/rCas9 can be used for functional analysis of miRNAs in rice.
Data presented in this study suggest that early isolation of T0 plants with biallelic homozygous and heteroallelic combinational mutations is a useful approach to obtain homogenous mutants in the next generation since the mutation patterns are stably inherited following the classic Mendelian law (Figure 5a). The generation of transgenic plants is one of the major limiting steps for crop research. In this study, we obtained 233 T0 lines with the biallelic mutation from 458 T0 transgenic plants (Table 1), indicating that at least 10 T0 plants should be generated to obtain five transgenic plants with biallelic mutations. The efficiency will increase up to 60.6% by using sgRNA with high GC ratio (Table 1). These results indicate that the use of rCas9 and sgRNA with a high GC ratio will greatly accelerate the generation of biallelic mutations on the target sequence in the T0 stage which is crucial for the efficient development of genome-edited rice. Further optimization of CRISPR/rCas9 in rice would promise higher mutation efficiency as well as the rapid creation of new varieties with valuable and novel traits.
4. Materials and Methods
4.1. Plasmid Construction
To apply the CRISPR/rCas9 system in rice, a recombinant codon-optimized Streptococcus pyogenes Cas9 for rice and single-guide RNA (sgRNA) were constructed in the pSB11 vector [35] through restriction-enzyme-mediated excision and ligation reactions. The rice codon-optimized Streptococcus pyogenes Cas9 was chemically synthesized by Bioneer in Korea (Daejeon, Korea). The expression of sgRNA and Cas9 was driven by the rice U6 promoter and Cauliflower mosaic virus 35S (CaMV 35S) promoter, respectively. Nuclear localization sequence (NLS) was fused to both N-terminus and C-terminus of the rice codon-optimized rCas9, and self-cleaving 2A peptide (P2A) and GFP were inserted into the C-terminal NLS sequence (Accession number MW296054) (Figure 1a). The 20 bp upstream sequence from the PAM (protospacer adjacent motif) sequence nearest to the miRNA seed sequence was used to guide RNA design.
For replacing gRNA in these vectors, the U6 promoter (pOsU6) and a sequence-specific gRNA were introduced into the pSB11 vector through HindIII and XbaI sites ((New England Biolabs, Ipswich, MA, USA)) (Figure 1a). To construct the pOsU6:sgRNA cassette containing a target-specific guide sequence, the guide RNA sequence was added at the 3′ and 5′ ends of the U6 promoter through three-step PCR reaction (Figure 1b). The final construct was sequenced to verify the correct insertion of the pOsU6:sgRNA-2×35S:rCas9 cassette. To generate OsmiR818b-overexpressing plants, the primary OsmiR818b sequence was amplified from rice (Oryza sativa L.ssp. japonica cv. Dongjin) total RNAs using the reverse transcription system (Promega, Madison, WI, USA) and PrimeSTAR HS DNA polymerase (Takara, Kyoto, Japan) with gene-specific primers (forward primer: CACCGATCGATCTCGTCGTCG, reverse primer: GAACCTTGCACATGACTTCAGCTAG). The amplified OsmiR818b sequence was cloned into the rice p700 transformation vector harboring GOS2 promoter for constitutive overexpression [36]. The final construct was named as GOS2::OsmiR818b. The primer information used for plasmid construction can be found in Table S1.
4.2. Transient Expression of CRISPR/rCas9 Using Protoplasts
The preparation of rice protoplasts was carried out as previously described [37]. For transient expression of sgRNA and CRISPR/rCas9, 50 μL of protoplasts (2–3 × 106 cells) was mixed with 15 μL of the CRISPR/rCas9 vector (1–2 μg) and 130 μL of 40% PEG solution (Sigma, St. Louis, MO, USA), and incubated for 15 min at 28 °C in the dark. After incubation, 1mL of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES (pH = 5.7)) was added into the mixture, and the protoplasts were incubated for 12 h at 28 °C. After incubation, the protoplasts were collected and used for genomic DNA extraction. The sgRNA target site was PCR-amplified and cloned into the pGEM-T Easy vector (Promega, USA). Colonies in the LB plates were picked and their insert sequences were determined by the Sanger method using target-specific primers (Cosmogenetech, Seoul, Korea).
4.3. Plant Transformation
Transgenic rice plants were obtained by A. tumefaciens strain LBA4404-mediated transformation of rice (Oryza sativa L. cv. Dongjin) embryonic callus as previously described [38]. The CRISPR/rCas9 or GOS2::OsmiR818b constructs were introduced into Agrobacterium tumefaciens strain LBA4404 by the triparental mating method with the helper cell.
4.4. Analysis of Mutation Frequency in Transgenic Plants by Sanger Sequencing
To analyze the mutation frequencies and spectrum of DNA modifications, target regions were amplified by PCR using gene-specific primers and genomic DNA extracted from leaves of non-transgenic and transgenic plants. Plant genomic DNA preparation was carried out using Qiagen DNeasy® 96 Plant Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. PCR reaction was performed to amplify the genomic regions including sgRNA-targeting sequence using the gene-specific primers (Table S1). PCR conditions were as follows: 98 °C for 2 min and 35 cycles of 98 °C for 30 sec, 60 °C for 10 sec, and 72 °C for 30 sec. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germany) and used for further direct sequencing and cloning into the pGEM-T Easy vector (Promega, USA).
4.5. miRNA Detection Using Stem–Loop RT-PCR
Total RNA was extracted from leaves using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following a previously-described protocol [39]. For analyzing the level of mature miRNAs, stem–loop reverse transcription followed by RT-PCR was performed as described previously [40,41]. Next, 200 ng of total RNA was treated with RNAase-free DNase I (Promega, USA), and used for first-strand synthesis reactions using gene-specific RT primers and the Superscript III Reverse transcriptase (Invitrogen, USA). Reactions were performed at 16 °C for 45 min, followed by 60 cycles of 30 °C for 45 s, 42 °C for 45 s, and 50 °C for 1 s, in a 20 μL mixture containing 50 U of Superscript III RT (Invitrogen, USA), 4 U of RNaseOUT (Invitrogen, USA), and 1 μM stem–loop RT primer. The products were used for quantification of mature miRNA through qRT-PCR analysis with miRNA-specific forward and universal reverse primers. The rice U6 small nuclear RNA (snRNA) gene was used as an RNA loading control. A list of primers used in these experiments is available in Table S1.
4.6. Drought-Stress Treatments and Tolerance Evaluation
Thirty independent OsmiR818b-overexpressing transgenic lines were produced, and plants that grew normally without stunting were selected to eliminate the effects of somaclonal variations. Then, copy numbers of the transgenic plants were determined by TaqMan Q-PCR (Thermo Fisher, Waltham, MA, USA) using proves specific for the bar gene. Three independent single copy homozygous transgenic plants were further selected based on expression level and antibiotic segregation analysis. Nineteen independent transgenic plants expressing CRISPR/rCas9 with the sgRNA targeting OsmiR818b were produced to generate CRISPR/Cas9miR818b mutants. The CRISPR/Cas9miR818b #19 that contained monoallelic mutation (24bp deletion) on OsmiR818b locus was selected at the T0 stage. The plants were propagated, and the biallelic homozygous mutant of OsmiR818b was obtained at the T1 stage. T2 seeds of the homozygous mutant that contained Cas9 T-DNA were used for drought treatments. Non-transgenic plants (O. Sativa cv. Dongjin), OsmiR818b-overexpressing (T3 generation), and CRISPR/Cas9miR818b (T2 stage)transgenic plants were sown on MS solid media and incubated in the dark growth chamber for two days at 28 °C. Two-day-old rice seedlings were transferred to the growth chamber with 16h light/ 8 h dark cycle for one additional day. Thirty seedlings from each line were transplanted into soil pots with a container and grown for an additional four weeks in a greenhouse (16h light/8h dark) at 30 °C. Drought stress was imposed by withholding water for 4 days. The plants were further incubated after re-watering for 6 days. Drought-induced symptoms were monitored by imaging transgenic and NT plants at the indicated time points using NEX-5N camera (Sony, Tôkyô, Japan).
Acknowledgments
We thank the Rural Development Administration and Kyungpook National University for providing the rice paddy fields.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/24/9606/s1. Table S1: The primer list used in this study. Figure S1: Determination of mutation types from PCR products. Figure S2: Analysis of CRISPR/rCas9-mediated IN/Del patterns in CRISPR/rCas9OsNAC14 transgenic plants.
Author Contributions
P.J.C., J.S.S., and J.-K.K. designed experiments; P.J.C., H.C., N.O., J.C., S.W.B., S.E.J., and H.J. performed experiments; P.J.C., J.S.S., and J.-K.K. wrote the manuscript and prepared the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the New Breeding Technologies Development Program (Project No. PJ01477201 to J.-K.K.), Rural Development Administration, Republic of Korea and the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (NRF-2013R1A6A3A04060627 to P.J.C. and 2018R1C1B6006927 to J.S.S.).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Gaj T., Gersbach C.A., Barbas C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra R., Joshi R.K., Zhao K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2020;18:20–31. doi: 10.1111/pbi.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J., Deng K., Cheng Y., Zhong Z., Tian L., Tang X., Tang A., Zheng X., Zhang T., Qi Y., et al. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 2017;8:1598. doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R., Marraffini L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A Programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H., Wu J.-J., Tang T., Liu K.-D., Dai C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-07871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruegmann T., Deecke K., Fladung M. Evaluating the efficiency of gRNAs in CRISPR/Cas9 mediated genome editing in poplars. Int. J. Mol. Sci. 2019;20:3623. doi: 10.3390/ijms20153623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Pan X., Cobb G.P., Anderson T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Iwakawa H.-O., Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 12.Wang J., Mei J., Ren G. Plant microRNAs: Biogenesis, homeostasis, and degradation. Front. Plant Sci. 2019;10:360. doi: 10.3389/fpls.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C., Li D., Mao D., Liu X., Ji C., Li X., Zhao X., Cheng Z., Chen C., Zhu L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.) Plant Cell Environ. 2013;36:2207–2218. doi: 10.1111/pce.12130. [DOI] [PubMed] [Google Scholar]
- 14.Djami-Tchatchou A.T., Sanan-Mishra N., Ntushelo K., Dubery I.A. Functional roles of microRNAs in agronomically important plants—Potential as targets for crop improvement and protection. Front. Plant Sci. 2017;8:378. doi: 10.3389/fpls.2017.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 16.Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X., Chen X., Tang G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–427. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichel M., Li Y., Li J., Millar A.A. Inhibiting plant microRNA activity: Molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J. 2015;13:915–926. doi: 10.1111/pbi.12327. [DOI] [PubMed] [Google Scholar]
- 19.Bi H., Fei Q., Li R., Liu B., Xia R., Char S.N., Meyers B.C., Yang B. Disruption of miRNA sequences by TALENs and CRISPR/Cas9 induces varied lengths of miRNA production. Plant Biotechnol. J. 2020;18:1526–1536. doi: 10.1111/pbi.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung P.J., Jung H., Jeong D.-H., Ha S.-H., Choi Y.D., Kim J.-K. Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genom. 2016;17:563. doi: 10.1186/s12864-016-2997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B., Yang X., Yang C., Li M., Guo Y. Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci. Rep. 2016;6:20315. doi: 10.1038/srep20315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Mikami M., Toki S., Endo M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 2015;88:561–572. doi: 10.1007/s11103-015-0342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson R.A., Gurevich V., Levy A.A. A rapid assay to quantify the cleavage efficiency of custom-designed nucleases in planta. Plant Mol. Biol. 2013;82:207–221. doi: 10.1007/s11103-013-0052-1. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., Mao Y., Yang L., Zhang H., Xu N., et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014;12:797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 26.Ren F., Ren C., Zhang Z., Duan W., Lecourieux D., Li S., Liang Z. Efficiency optimization of CRISPR/Cas9-mediated targeted mutagenesis in grape. Front. Plant Sci. 2019;10:612. doi: 10.3389/fpls.2019.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan Q., Wang Y., Chen K., Liang Z., Li J., Zhang Y., Zhang K., Liu J., Voytas D.F., Zheng X., et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant. 2013;6:1365–1368. doi: 10.1093/mp/sss162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basso M.F., Ferreira P.C.G., Kobayashi A.K., Harmon F.G., Nepomuceno A.L., Molinari H.B.C., Grossi-de-Sa M.F. MicroRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechol. J. 2019;17:1482–1500. doi: 10.1111/pbi.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang H., Yi B., Ma R., Zhang X., Zhao H., Xi Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 2016;6:22312. doi: 10.1038/srep22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y., Dai Z., Liang Y., Yin M., Ma K., He M., Ouyang H., Teng C.-B. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 2014;4:3943. doi: 10.1038/srep03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurata J.S., Lin R.-J. MicroRNA-focused CRISPR-Cas9 library screen reveals fitness-associated miRNAs. RNA. 2018;24:966–981. doi: 10.1261/rna.066282.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aquino-Jarquin G. Emerging Role of CRISPR/Cas9 Technology for MicroRNAs Editing in Cancer Research. Cancer Res. 2017;77:6812–6817. doi: 10.1158/0008-5472.CAN-17-2142. [DOI] [PubMed] [Google Scholar]
- 35.Komari T., Hiei Y., Saito Y., Murai N., Kumashiro T. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 1996;10:165–174. doi: 10.1046/j.1365-313X.1996.10010165.x. [DOI] [PubMed] [Google Scholar]
- 36.Jeong J.S., Kim Y.S., Baek K.H., Jung H., Ha S.H., Do Choi Y., Kim M., Reuzeau C., Kim J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shim J.S., Oh N., Chung P.J., Kim Y.S., Choi Y.D., Kim J.K. Overexpression of OsNAC14 Improves Drought Tolerance in Rice. Front. Plant Sci. 2018;9:310. doi: 10.3389/fpls.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang I.-C., Nahm B.H., Kim J.-K. Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol. Breed. 1999;5:453–461. doi: 10.1023/A:1009665314850. [DOI] [Google Scholar]
- 39.Chung P.J., Park B.S., Wang H., Liu J., Jang I.-C., Chua N.-H. Light-inducible MiR163 targets PXMT1 transcripts to promote seed germination and primary root elongation in Arabidopsis. Plant Physiol. 2016;170:1772–1782. doi: 10.1104/pp.15.01188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.