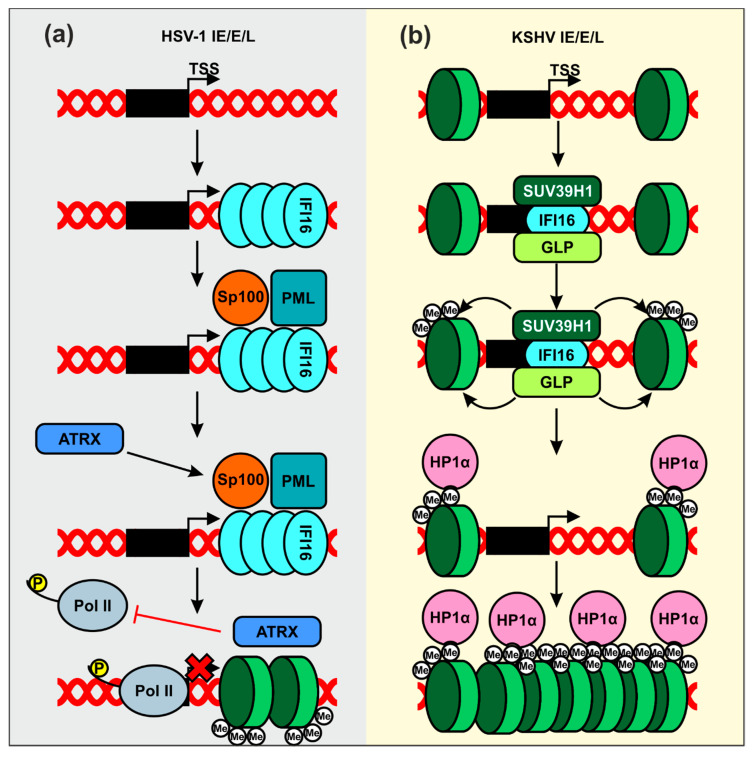
Figure 3.
Proposed mechanisms of IFI16-mediated inhibition of herpesviral gene expression by modification of chromatin. (a) IFI16 recognizes HSV-1-derived DNA and assembles filamentous structures, which allows recruitment of transcriptional repressors such as ATRX. This reduces the association of elongation-competent RNA Pol II on immediate-early (IE), early (E), and late (L) genes and further leads to chromatinization of viral genes (green discs indicate H3K9). (b) IFI16 complexed with SUV39H1 and G9a-like protein (GLP) binds to the viral DNA, and the latter two promote the first methylation event on existing histones. Trimethylated H3K9 is recognized by HP1α, which recruits additional chromatin-modifying enzymes that further compact the viral genes into a repressed state.