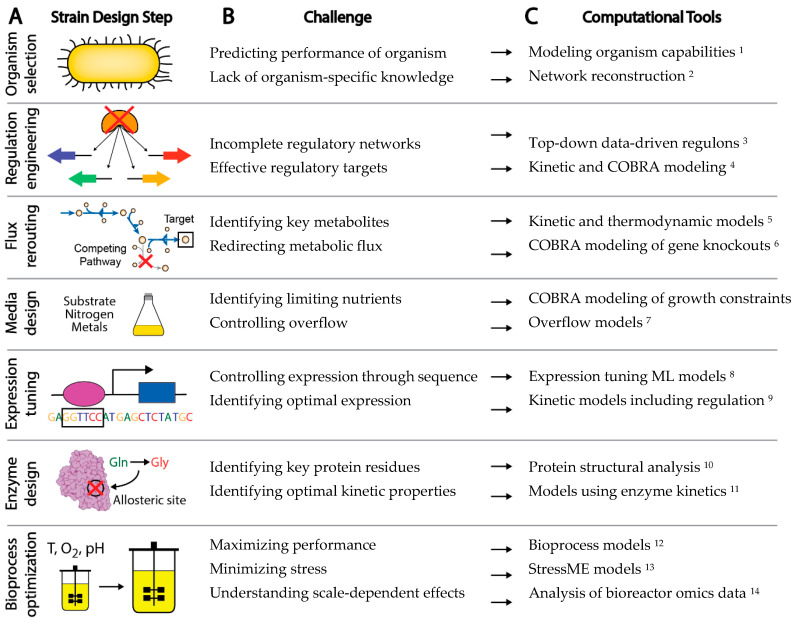
Figure 1.
Challenges and computational solutions for a typical strain design workflow. (A) Typical experimental steps in the development of a new strain design. (B) Common challenges encountered at each strain design step. (C) Computational tools that may be used to meet the strain design challenges. Note that the design steps, challenges, and computational tools highlighted here are intended to be exemplative rather than comprehensive. 1, Modeling organism capabilities [27]; 2, Network reconstruction [18]; 3, Top-down data-driven regulons [28]; 4, Kinetic and COBRA modeling [26]; 5, Kinetic and thermodynamic models [29]; 6, COBRA modeling of gene knockouts [30]; 7, Overflow models [31]; 8, Expression tuning ML models [32]; 9, Kinetic models including regulation [33]; 10, Protein structural analysis [34]; 11, Models using enzyme kinetics [35]; 12, Bioprocess models [36]; 13, StressME models [37,38,39]; 14, Analysis of bioreactor omics data [40].