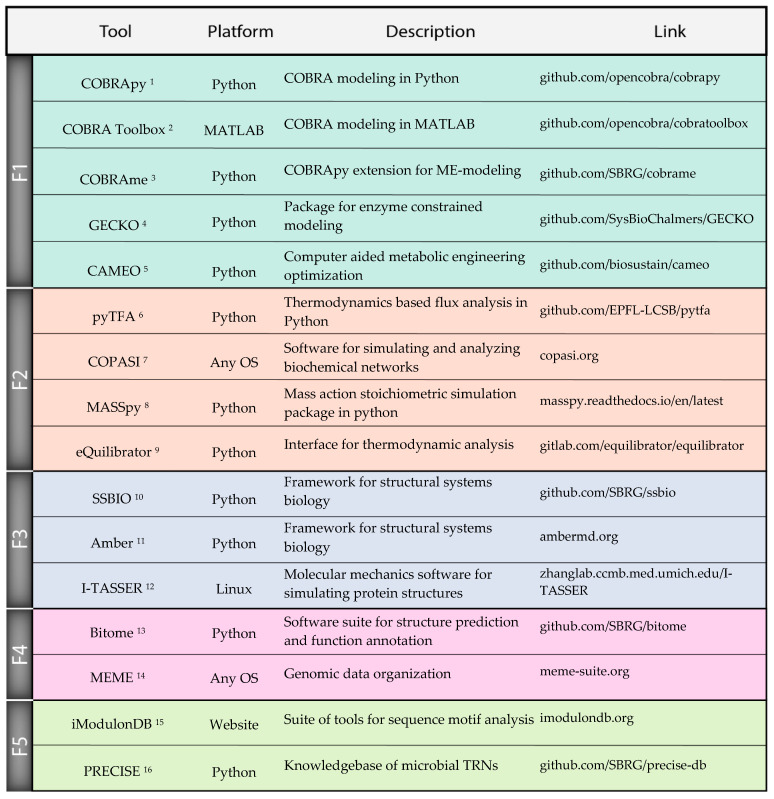
Figure 3.
A selection of actively maintained software for computational design and analysis of microbial phenotypes. We focus on Python tools due to the popularity of the language as well as potential for integration in a single strain design workflow, but also include important packages in other languages and standalone applications. Frontier 1: Packages for constraint-based reconstruction and modeling, proteome allocation modeling, and strain design optimization. Frontier 2: Kinetics and thermodynamics packages for model parameterization, simulation, and thermodynamics constrained modeling. Frontier 3: Software for annotating and visualizing structures as well as integrating 3D structural information with systems biology approaches. Frontier 4: Python package for storing, organizing, and analyzing genome sequences. Frontier 5: Online knowledgebase and software for determining transcriptional regulatory networks using ICA decomposition methods. 1 COBRApy [42]; 2 COBRA Toolbox [17]; 3 COBRAme [20]; 4 GECKO [52]; 5 CAMEO [43]; 6 pyTFA [95]; 7 COPASI [96]; 8 MASSpy [94]; 9 eQuilibrator [89]; 10 SSBIO [106]; 11 Amber [111]; 12 I-TASSER [107]; 13 Bitome [120]; 14 MEME [117]; 15 iModulonDB [125]; 16 PRECISE [28].