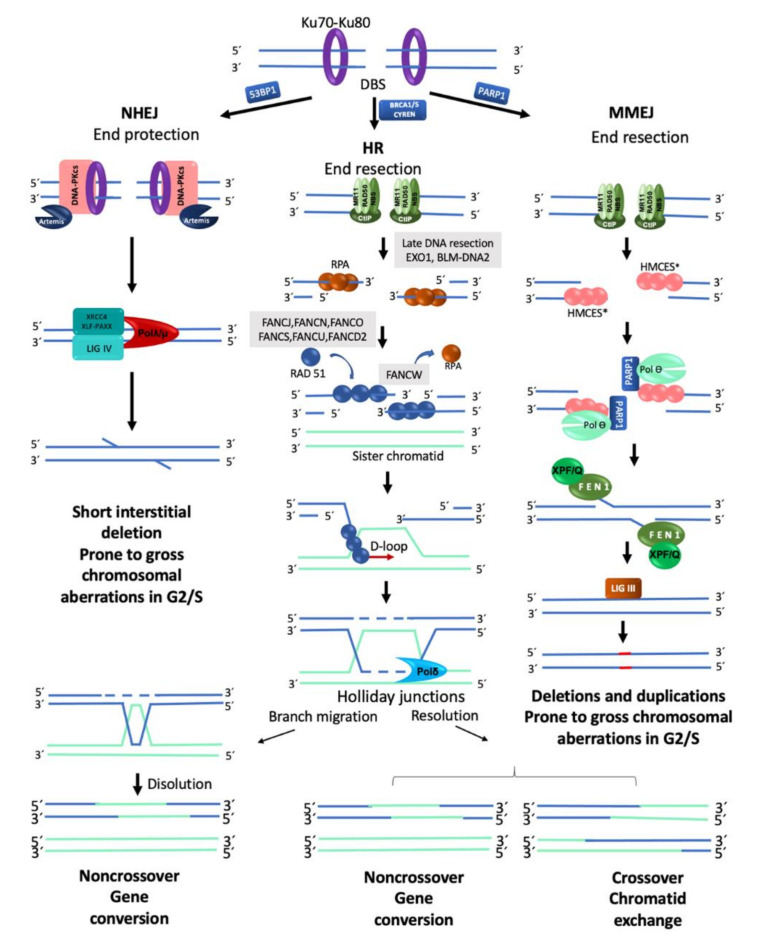
Figure 2.
Major mechanisms of double-strand break repair. NHEJ, Non-Homologous End Joining. DNA ends are protected by KU70/KU80, which prevents their end-processing. When the ends are incompatible, a segment of up to four nucleotides is detected, Artemis eliminates the remaining incompatible segments and ligation is carried out by LIGIV-XRCC4. This pathway can repair a DSB without chromosome modification; however, during S/G2 and in the presence of several DSBs, it is considered prone to generate chromosomal alterations. HR, Homologous Recombination. This pathway is only available during the S/G2 phases of the cell cycle since it requires a homologous template; HR is the best choice to maintain sequence fidelity because in general it repairs in an error-free manner. A ssDNA 3′ overhang is produced by the action of the MRN-CtIP complex; it is first covered by RPA proteins which are later replaced by RAD51 to form the nucleoprotein strand that will carry out the invasion of the DNA of the sister chromatid in order to use it as a template to restore the continuity of the original nucleotide sequence. MMEJ, Microhomology-Mediated End Joining. PARP1 prevents KU70/KU80 binding to DNA ends and allows the recruitment of MRN-CtIP to initiate end-resection, creating a short 3′ overhang. This overhang is preferentially covered by HMCES which channels the damage to be repaired by MMEJ instead of HR. PARP1-POLQ search for microhomology of 2–20 bp and align the strands. The resulting flaps are eliminated by XPF/FANCQ-FEN1. Alternatively, POLQ can direct DNA synthesis to add nucleotides to make DNA ends compatible; this end processing generates in situ deletions and duplications. In addition, if this pathway is active during S/G2, and several DSBs coincide in time and space, gross chromosomal aberrations are formed because, similar to NHEJ, they do not require long stretches of homology to ligate the DNA ends [23,26]. * RPA or HMCES (5-hydroxymethylcytosine binding, embryonic stem cell-specific protein) [26,28].