FIGURE 3.
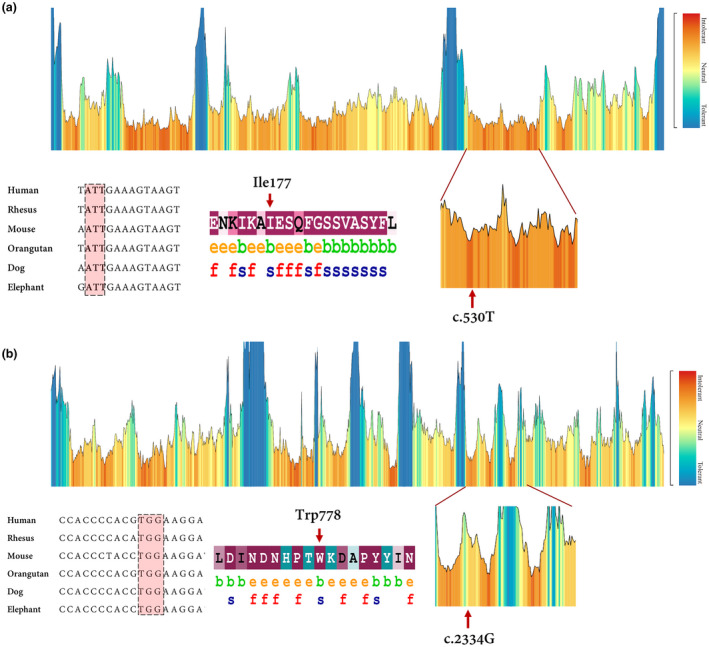
(a) the tolerance landscape depicts a missense over the synonymous ratio calculated as a sliding window over the entirety of the protein. The missense variation is annotated from the gnomAD data set and the landscape provides some indication of regions that are intolerant to missense variation. In this TMC1 tolerance landscape, the region harboring the novel missense variant can be seen as intolerant if compared with other parts in this protein. Nucleotide alignment showing high conservation of the codon residue which encodes Ile 177. The ConSurf server was applied to estimate conservation scores for the amino acid residue substituted by the missense variant. Scores ranged from 1 to 9, where a score of 9 represented a highly conserved residue (Glaser et al.,2003). ConSurf demonstrates evolutionary conservation profiles for proteins of unknown/known structure in the PDB according to the phylogenetic relations. (b) MetaDome database was used to identify the intolerant regions (surrounding the c.2334G>A variant). As depicted, the novel variant is located in a highly intolerant region. Data derived from nucleotide alignment and ConSurf show that the c.2334G or Trp778 is highly conserved
