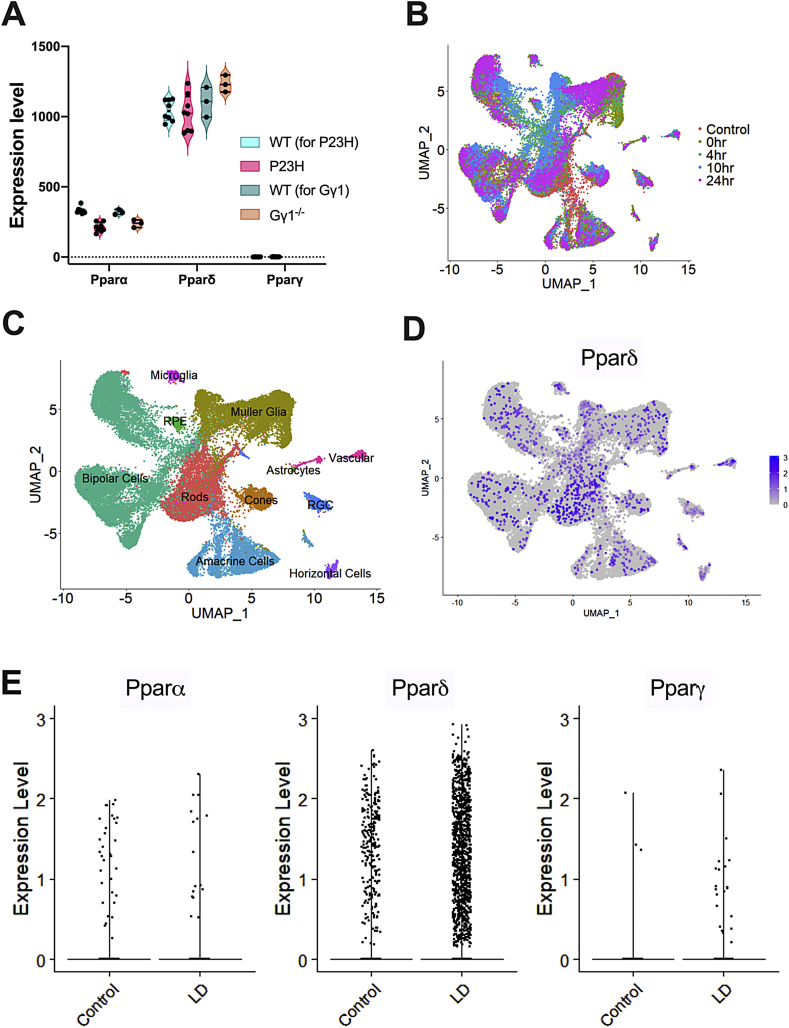
Fig. 1.
RNAseq and single-cell RNAseq data analysis for expression of Pparα, Pparδ, and Pparγ in the retina. (A) Violin plots showing gene expression levels of Pparα, Pparδ, and Pparγ in wildtype (WT), heterozygous RhoP23H knock-in (P23H), and transducin gamma knockout (Gγ1−/−) mice from whole-retina RNA-seq data. Each dot represents a single sequencing library from whole retinas, and are biological replicates. DESeq2 and the Wald test was used to determine differentially expressed genes in the dataset. * Indicates statistical significance, as compared to the WT for each group. The adjusted P-values were 1.74E-13 for P23H and 0.0031 for Gγ1−/−. B) Uniform manifold approximation and projection (UMAP) plots of retinal cells in control and light damaged (LD) retinas. Each dot represents a single cell and the colors indicate the time point after LD. Each library consisted of a male and female retina that was subjected to its specified treatment. Each sample group had 2 replicates, except for the 24-h post-light damage group. Cells are clustered by differentially expressed genes. (C) UMAP plot of retinal cell clusters following LD representing all of the retinal cell types. Cells in each cluster were identified based on the expression of known cell markers. Colors represent different cell types. (D) UMAP plot of single cell RNA sequencing (scRNAseq) data with Pparδ-expressing cells labeled in purple. (E) Scatter plots showing gene expression levels of Pparα, Pparδ, and Pparγ in scRNAseq data from WT and light-damaged retinas. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)