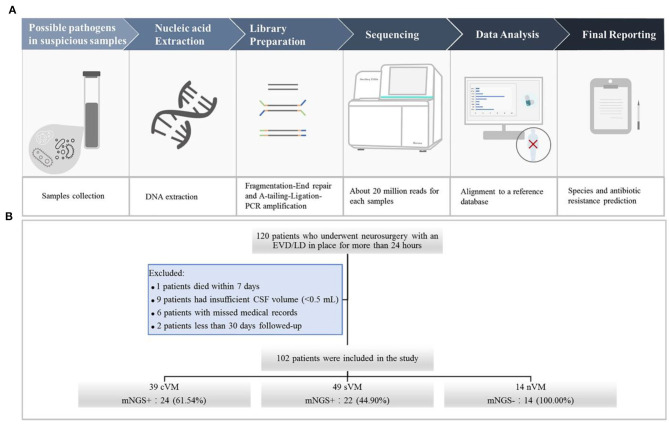
Figure 1.
(A) Schematic of the mNGS workflow. After collecting CSF samples, DNA was extracted using the TIANamp Magnetic DNA Kit (Tiangen). DNA libraries were prepared using the KAPA Hyper Prep kit (KAPA Biosystems) and fragmentation-end repair and a-tail-ligation PCR amplification. Libraries were quantified, pooled, and loaded onto the sequencer. Using an in-house-developed bioinformatics pipeline, high-quality sequencing data were obtained. Reads were aligned to the NCBI microorganism genome database for pathogen identification. Finally, we reported the results of mNGS according to the criteria. (B) Flowchart of study participants.