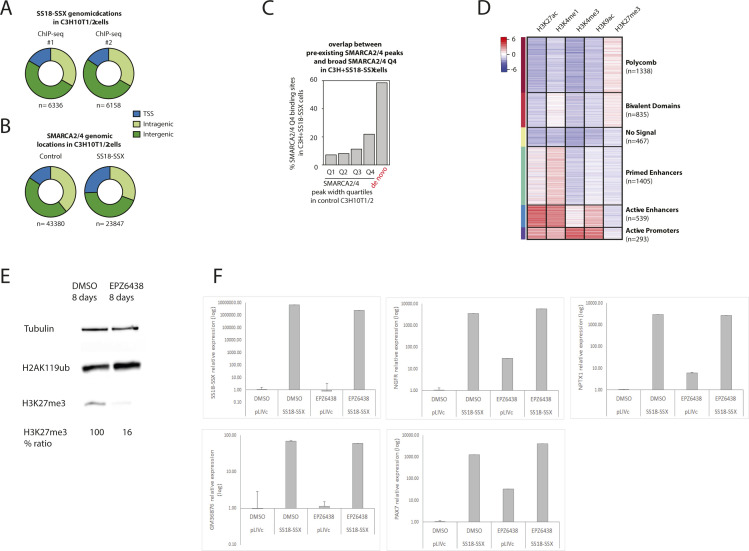
Figure S4. Related to Fig 4. SS18-SSX chromatin binding and activity are independent of preexisting H3K27me3.
(A) Pie charts showing genomic locations of SS18-SSX–binding sites in C3H10T1/2 cells stably expressing SS18-SSX in two independent ChIP-seq experiments. (B) Pie charts showing genomic locations of SMARCA2/4–binding sites in control and SS18-SSX–expressing C3H10T1/2 cells. (C) Barplot showing the percentage of preexisting and de novo SMARCA2 domains in each width quartile overlapping with SS18-SSX–binding sites in C3H10T1/2 cells transduced with SS18-SSX. (D) Heat map showing initial ChIP-seq signals for the indicated histone modifications in control cells at SS18-SSX–binding sites. The indicated categories were defined by hierarchical clustering and manually annotated based on chromatin states. (E) Western blot analysis of H3K27me3 and H2AK119ub expression levels in control (DMSO) and EPZ6438-treated C3H10T1/2 cells. Densitometric analysis, using ImageJ software package, is reported. (F) qPCR analysis of a panel of SS18-SSX target genes in control (DMSO) and EPZ6438-treated C3H10T1/2 cells infected with either Empty (pLIVc) or SS18-SSX–expressing lentiviral vectors. Error bars represent the SD of triplicate tests. Relative expression is reported on a logarithmic scale.