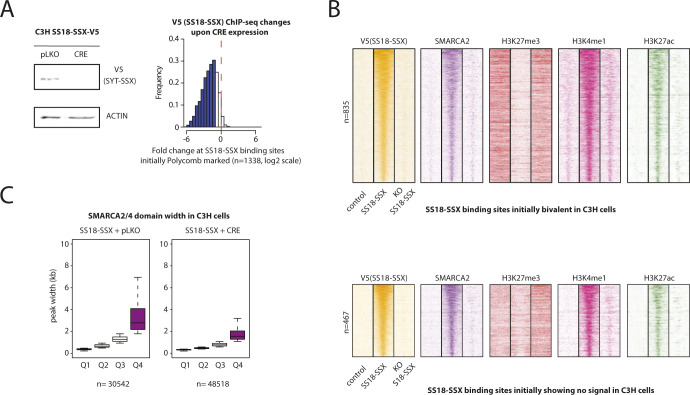
Figure S5. Related to Figs 4 and 5. Reversibility of the chromatin remodeling events induced by SS18-SSX in mouse C3H10T1/2 cells.
(A) Left: Immunoblot showing loss of V5-SS18-SSX in C3H10T1/2 cells upon CRE expression. Right: Histogram showing changes in V5 (SS18-SSX) ChIP-seq signals upon SS18-SSX removal in C3H10T1/2 cells expressing SS18-SSX at binding sites that bore the polycomb mark in control cells (n = 1,338 sites identified). (B) Heat maps showing ChIP-seq signals for V5 (SS18-SSX), SMARCA2/4, H3K27me3, H3K4me1, and H3K27ac at SS18-SSX–binding sites initially identified in control cells as bivalent or without signals (Fig 4E). 20-kb windows centered on SS18-SSX–binding sites are shown. (C) Boxplot showing the distribution of peak widths for SMARCA2/4 in C3H10T1/2 cells expressing SS18-SSX upon control pLKO vector (left) or CRE expression (right) per quartile.