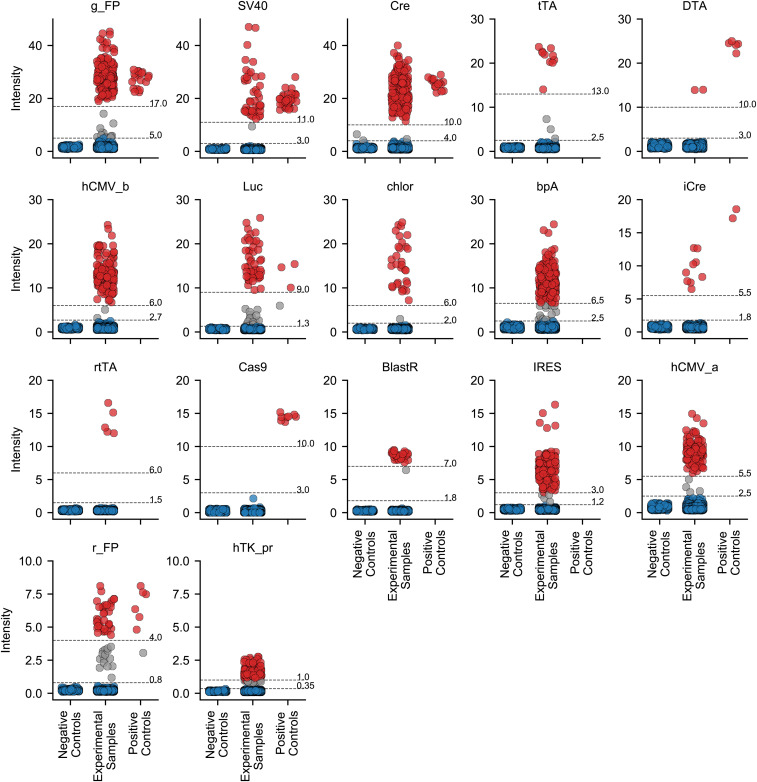
Figure 9.
Detection of genetic constructs validated in MiniMUGA. For each construct, samples are shown as dots and classified as negative controls (left), experimental (center), and positive controls (right). The dot color denotes whether the sample is determined to be negative (blue), positive (red), or questionable (gray) for the respective construct. For each construct, the gray horizontal lines represent data-driven ad hoc thresholds discriminating between presence and absence. Note for each construct, the y-axis scale is different. MUGA, Mouse Universal Genotyping Array; g_FP; ‘greenish’ fluorescent protein; SV40; SV40 large T antigen; Cre, Cre recombinase; tTA, tetracycline repressor protein;; DTA, Diptheria toxin; hCMV_b, Human CMV enhancer version b; Luc, Luciferase and firefly luciferase; chloR, Chloramphenicol acetyltransferase; bpA, Bovine growth hormone poly A signal sequence; iCre, iCre recombinanse; rtTA, Reverse improved tetracycline-controlled transactivator; cas9, CRISPR associated protein 9; BlastR, Blasticidin resistance; IRES, Internal Ribosome Entry Site; hCMV_a, hCMV enhancer version a; r_FP, ‘reddish’ fluorescent protein; hTK_pr, Herpesvirus TK promoter.