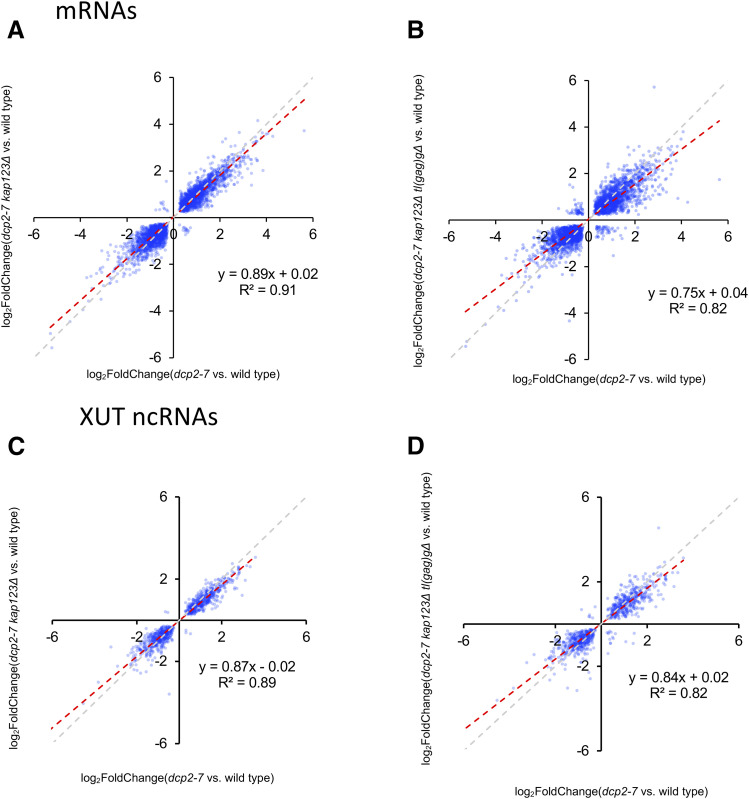
Figure 10.
Suppressors ameliorate defects in expression of protein coding mRNA and XUT noncoding RNA in dcp2-7 mutant. (A and B) Defective mRNA expression in dcp2-7 mutant was alleviated by kap123∆ and further by kap123∆ tl(gag)g∆. Plotted is the differential expression of mRNA loci in suppressor mutants vs. dcp2-7 mutant (relative to wild type). Of 6691 mRNA genes in S. cerevisiae, 3236 and 3302 transcripts that are statistically significant (adjusted P-value < 0.05 of DESeq2) were plotted, respectively. YBR115C, YCR097W, YEL021W, YNL145W, YER110C, and YPL187W are not plotted. (C and D) Defective XUT expression in dcp2-7 mutant was alleviated by kap123∆, and further by kap123∆ tl(gag)g∆. Plotted is the differential expression of XUT loci in suppressor mutants vs. dcp2-7 mutant relative to wild type. Of 1658 XUT genes in S. cerevisiae, 948 and 898 transcripts that are statistically significant (adjusted P-value < 0.05) were plotted, respectively. (A–D) The gray dashed line with slope 1 is the predicted outcome if kap123∆ and tl(gag)g∆ had no effect. The red dashed line with slope < 1 depicts linear regression analysis results.