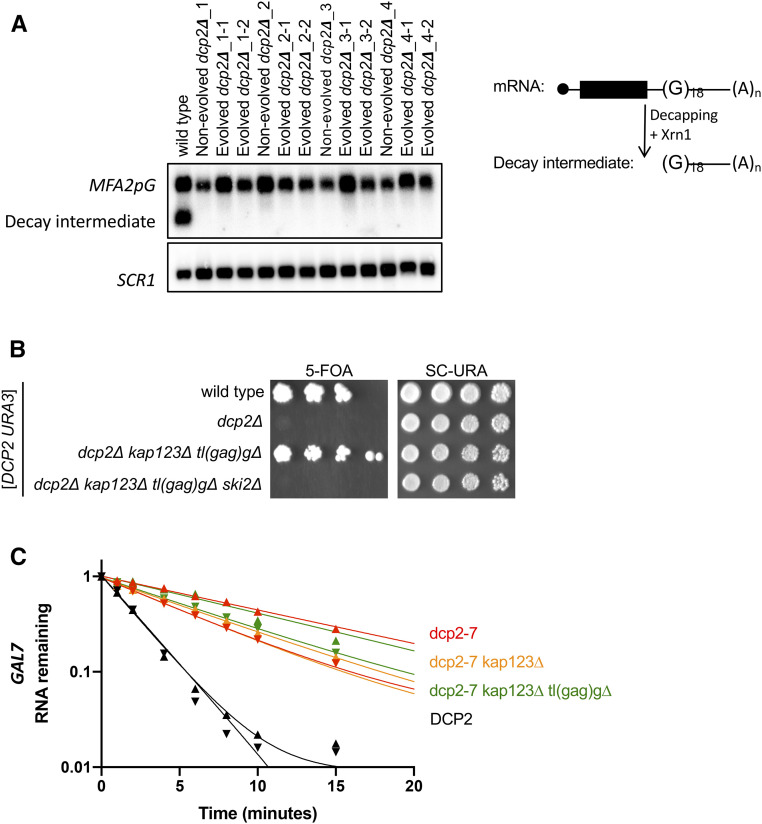
Figure 7.
Suppressor mutations of dcp2∆ do not restore mRNA decay. (A) 5′ to 3′ mRNA decay is defective in evolved dcp2∆ strains. Each strain was transformed with an MFA2pG reporter (right). Decapping, or endonucleolytic cleavage, followed by 5′ exonucleolytic decay of MFA2pG produces a decay intermediate that is a sensitive measure of decapping. Total RNA was isolated, and MFA2pG mRNA was analyzed by Northern blotting. A representative blot from three replicates is shown. Top panel was probed for MFA2pG, and the bottom panel for the loading control SCR1. Quantitation of all three replicates is in Figure S7. (B) RNA exosome-mediated decay is required in dcp2∆ suppressor mutants. dcp2∆ kap123∆ tl(gag)g∆ is no longer viable in the absence of SKI2. Each strain was made by sporulation of a quadruple heterozygous diploid transformed with a URA3 plasmid expressing DCP2. Haploid progeny were serially diluted and spotted on 5FOA and SC-URA (control) solid media. Shown is the growth of a representative haploid strain at day 5. (C) The stability of GAL7 mRNA is unaffected by kap123∆ and/or tl(gag)g∆ in DCP2-deficient cells. Cells exponentially growing at 21° in galactose were transferred to 37° for 1 hr. Transcription of GAL7 was repressed by the addition of dextrose at time 0, and cells were harvested at multiple time points. Total RNA was isolated and GAL7 mRNA levels were analyzed by Northern blotting. Plotted is remaining GAL7 mRNA levels relative to SCR1 levels of two biological replicates. Data point triangles are pointing up for one replicate, and down for the other.