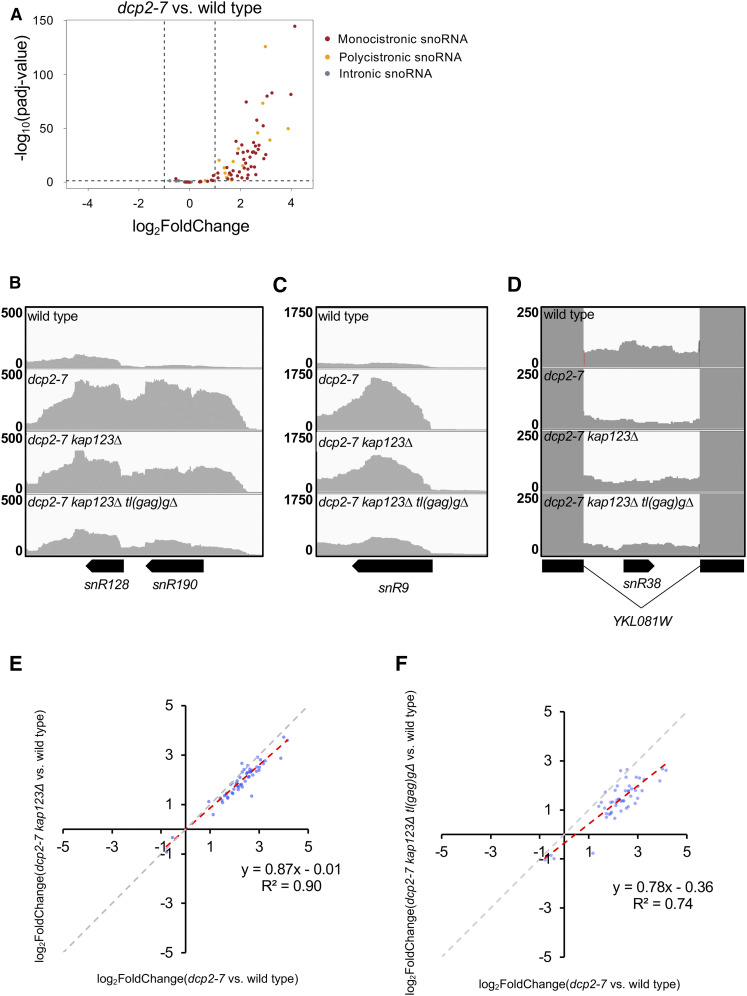
Figure 9.
Suppressors ameliorate expression defects from snoRNA loci in dcp2-7 mutant. (A) The expression of mono- and polycistronic snoRNA loci are upregulated in dcp2-7 mutant. A volcano plot showing differential expression of snoRNA genes in dcp2-7 strain vs. wild type. Monocistronic snoRNA are in red, polycistronic snoRNA genes are in orange, and intronic snoRNA genes are in gray. Dashed lines represent cut-off values for a twofold change in gene expression, and adjusted P-value < 0.05. (B–D) Upregulation of mono- and polycistronic snoRNA loci in dcp2-7 mutant were reduced by kap123∆, and further by kap123∆ tl(gag)g∆. RNA-seq read coverage for the snoRNA gene loci snR128-snR190 (dicistronic), snR9 (monocistronic), and snR38 (intronic) are shown. (E and F) Defective snoRNA expression in dcp2-7 mutant was alleviated by kap123∆ and further by kap123∆ tl(gag)g∆. Plotted is the differential expression of snoRNA loci in suppressor mutants vs. dcp2-7 mutant (relative to wild type). Of 77 snoRNA genes in S. cerevisiae, 53 and 49 transcripts that are statistically significant (adjusted P-value < 0.05 of DESeq2) were plotted, respectively. The gray dashed line with slope 1 is the predicted outcome if kap123∆ and tl(gag)g∆ had no effect. The red dashed line with slope < 1 depicts linear regression analysis results.