Figure 6:
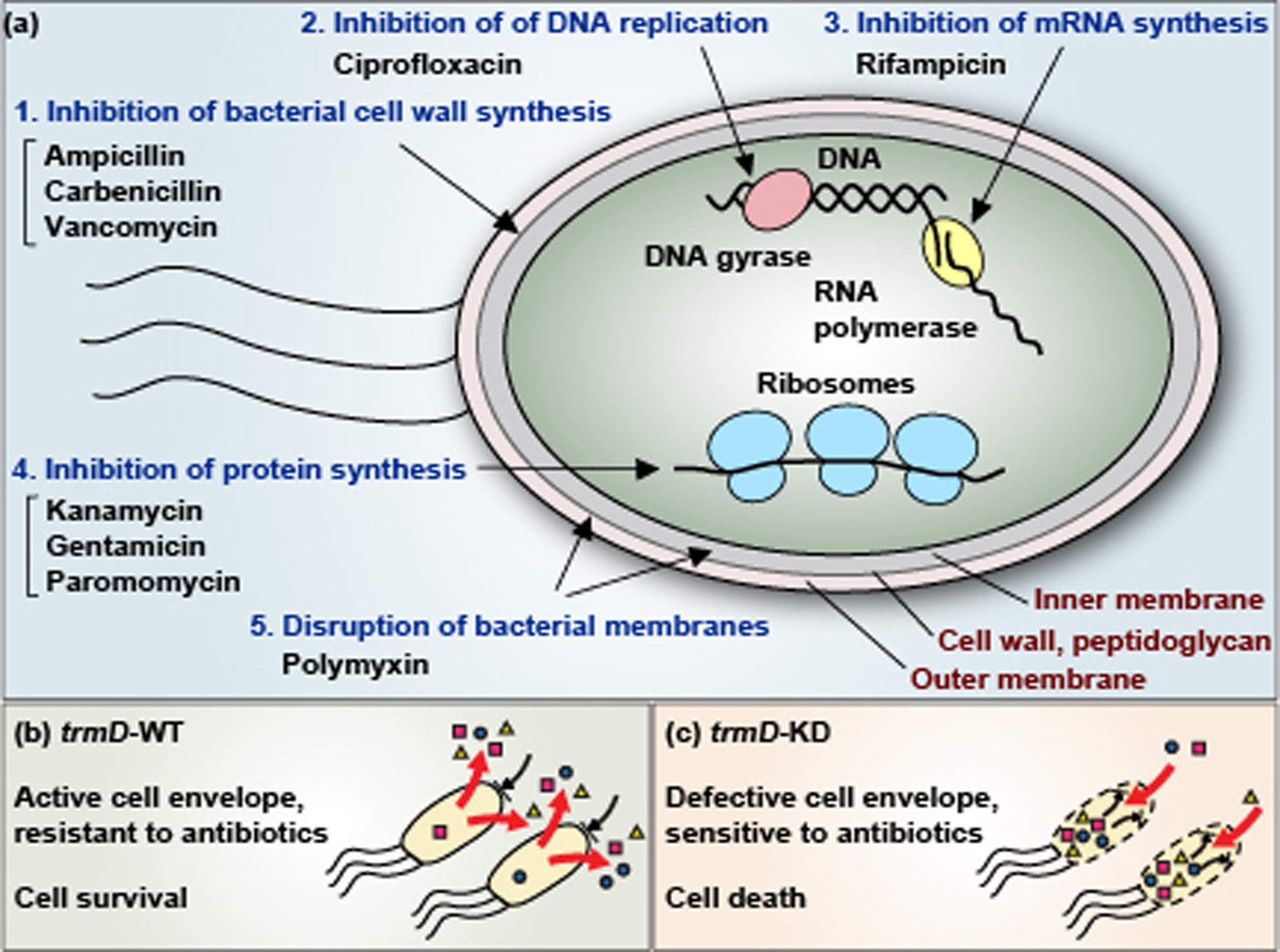
Control of biosynthesis of Gram-negative cell envelope by the TrmD-catalyzed m1G37-tRNA. (a) Mechanisms of action of antibiotics that become more potent in E. coli and Salmonella cells deficient of TrmD. These antibiotics are categorized into 5 mechanisms: (1) inhibition of bacterial cell wall synthesis, (2) inhibition of DNA replication, (3) inhibition of mRNA synthesis, (4) inhibition of protein synthesis on the ribosome, and (5) disruption of bacterial membranes. In all cases, the MIC value of each antibiotic decreases in trmD-KD cells relative to trmD-WT cells. (b) E. coli and Salmonella trmD-WT cells have an active cell envelope structure, due to the ability to synthesize m1G37-tRNA for translation of membrane-associated genes containing Pro codons, and are thus resistant to antibiotics via the action of the membrane permeability barrier and efflux pumps. (c) E. coli and Salmonella trmD-KD cells have a defective cell envelope, due to the deficiency of m1G37 and the inability to translate membrane-associated genes containing Pro codons, and are thus sensitive to antibiotic killing.
