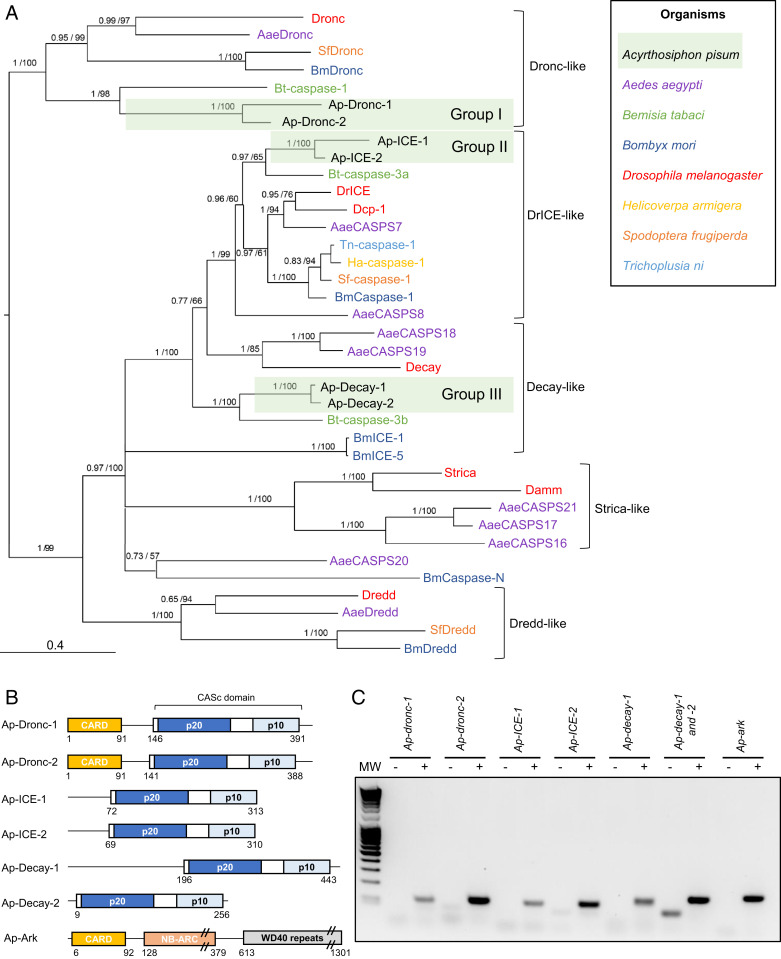
Fig. 1.
Identification of caspase- and adaptor protein-encoding genes in the pea aphid genome. (A) The phylogenetic relationships between caspase protein sequences found in Acyrthosiphon pisum and a selection of insect species based on CASc domain alignment. For each node, Bayesian posterior probability and bootstrap values are indicated. A group of putative A. pisum caspase paralogs are also indicated. Midpoint rooting was used to present the tree. (B) The domain architecture of the pea aphid putative caspases and adaptor proteins. Each caspase possesses a CASc domain (CASpase catalytic domain) composed of two distinct subunits (p20 and p10) and preceded by a prodomain of variable size. Ap-Dronc-1 and Ap-Dronc-2 possess an additional CARD domain (Caspase Recruitment Domain) at their N terminus. The putative adaptor protein Ap-Ark contains a CARD domain, a NB-ARC domain, and multiple WD40 repeats. The position of amino acids that mark the beginning and the end of the different domains are indicated below each structure. (C) Pea aphid putative caspases and adaptor proteins genes are expressed. The sequences were successfully amplified from a pool of cDNA obtained from whole aphids at different life stages. For each primer set, a negative control was performed on samples devoid of cDNA. MW, molecular weight marker.