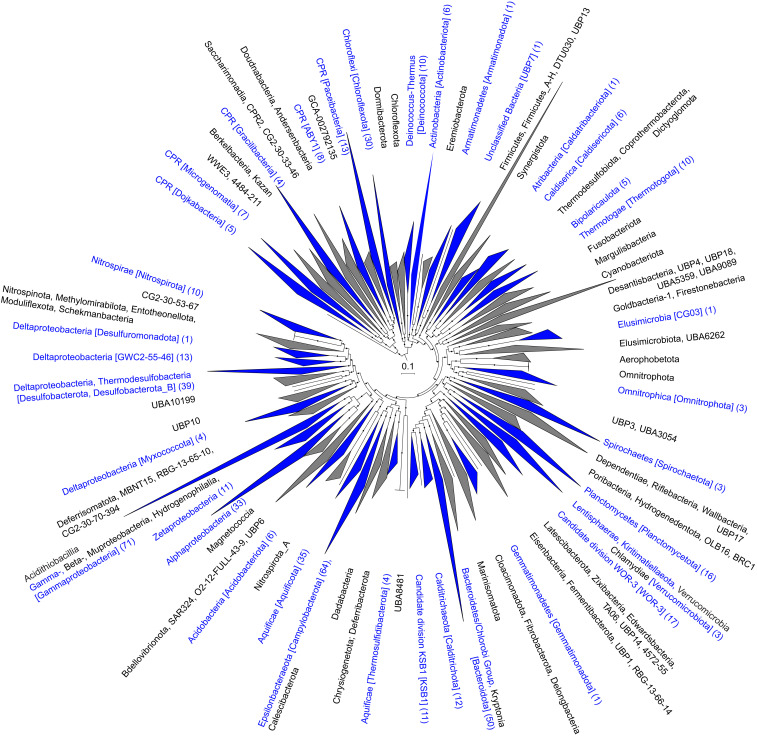
Fig. 4.
Maximum-likelihood phylogenetic reconstruction of bacterial metagenome-assembled genomes from Brothers volcano using GTDB-Tk. The tree was constructed using 120 bacterial marker genes. Branch support was determined with the SH test, and support values from 0.8 to 1.0 are indicated with filled circles. Collapsed taxonomic clades are shown as triangles, with blue triangles indicating clades containing Brothers volcano MAGs with the number of MAGs shown in parentheses. Clades with Brothers MAG are labeled using NCBI taxonomy followed by GTDB-Tk taxonomy in brackets, while gray clades without Brothers MAGs are shown with GTDB-Tk taxonomy only. Due to the extensive reordering of the Tree of Life within GTDB-Tk taxonomy, the NCBI taxonomy is approximate and represents large-scale taxa equivalencies. The scale bar indicates expected amino acid substitutions per site. The uncollapsed phylogenetic tree used to create this figure is available online (28) at https://itol.embl.de/shared/alrlab and on FigShare in phyloXML format (21) at https://doi.org/10.6084/m9.figshare.c.5099348.