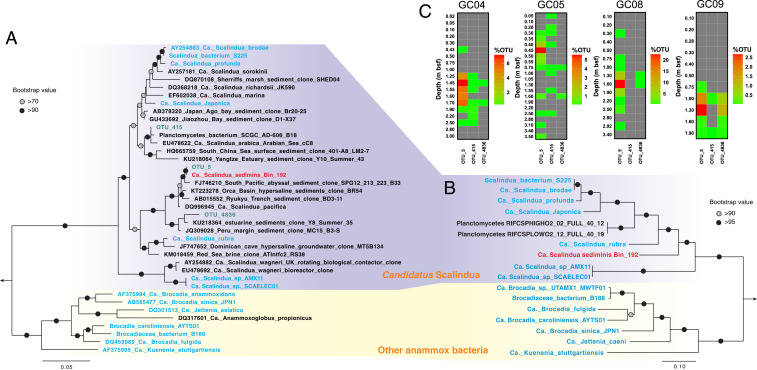
Fig. 3.
Phylogeny and vertical distribution pattern of Scalindua bacteria in AMOR sediments. (A) Maximum-likelihood phylogenetic tree of 16S rRNA genes of anammox bacteria. Sequences of the three Scalindua OTUs recovered from the sediments via 16S rRNA gene amplicon sequencing are shown in green. (B) Maximum-likelihood phylogenetic tree of anammox bacteria inferred from 14 concatenated ribosomal proteins. In both A and B, Ca. S. sediminis is highlighted in red, while other anammox bacteria are highlighted in green. Paludisphaera borealis PX4 and Isosphaera pallida were used as the outgroup for both trees. Bootstrap values higher than 70 and 90 are shown on nodes with open and filled circles, respectively. The scale bars correspond to estimated substitution per site. (C) Distribution of Scalindua OTUs in the four sediment cores. Depths of sediment horizons (meters below seafloor) are indicated on the vertical axis for each core. Note different scales are used for the anammox OTU percentages (of total community) between cores.