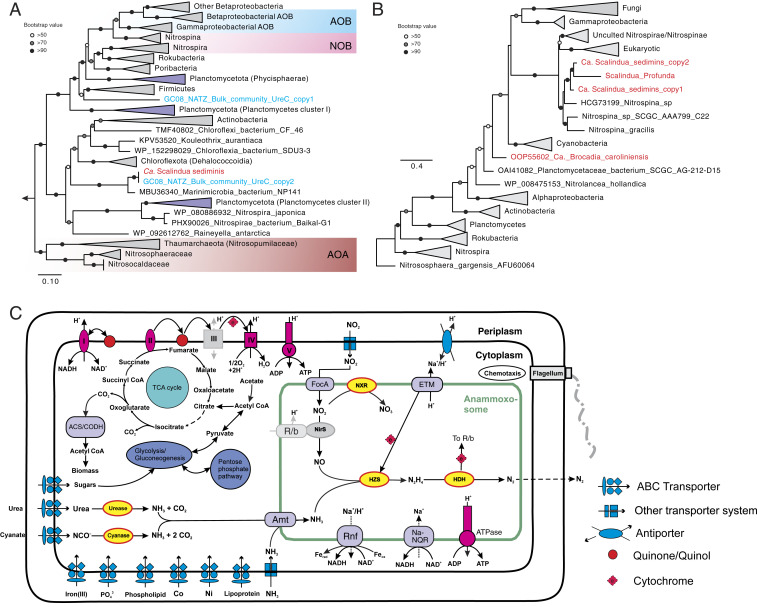
Fig. 4.
Metabolic potential of “Ca. S. sediminis”. (A) Maximum-likelihood phylogenetic tree of UreC (the catalytic alpha subunit of urease). The two UreC sequences detected in the metagenome assembly before binning are shown in blue and the one of Ca. S. sediminis is highlighted in red. Clades of nitrifying groups (i.e., AOA, AOB, and NOB) are highlighted in the shaded boxes, while sequences and clades of sequences of other Planctomycetes are shown in light purple. Bootstrap values of >50 are shown with symbols listed in the legend. The scale bar shows estimated sequence substitutions per residue. (B) Maximum-likelihood phylogenetic tree of cyanate hydratase (cynS) amino acid sequences (∼150 aa). The two copies of CynS detected in the Ca. S. sediminis genome and other anammox bacteria are highlighted in red. (C) Reconstruction of cell metabolic pathways based on the “Ca. S. sediminis” genome annotation. Enzyme complexes of the electron transport chain are labeled with Roman numerals. The flow of electron transfer is represented by blue arrows. HZS, hydrazine synthase; HDH, hydrazine dehydrogenase; HAO, hydroxylamine oxidase; NXR, nitrite oxidoreductase; TCA cycle, tricarboxylic acid cycle; Amt, ammonium transporter; ETM, electron transport module; FocA, nitrite/formate transporter; Rnf, ferredoxin-NAD:oxidoreductase; ACS/CODH, Acetyl-CoA synthase/carbon monoxide dehydrogenase, or Wood–Ljungdahl pathway; Na-NQR, Na-translocating NADH-quinone. Modules not detected in the genome annotation are shown in gray. Not drawn to scale. Uncertainties also exist for the position and size of the anammoxosome in the cells.