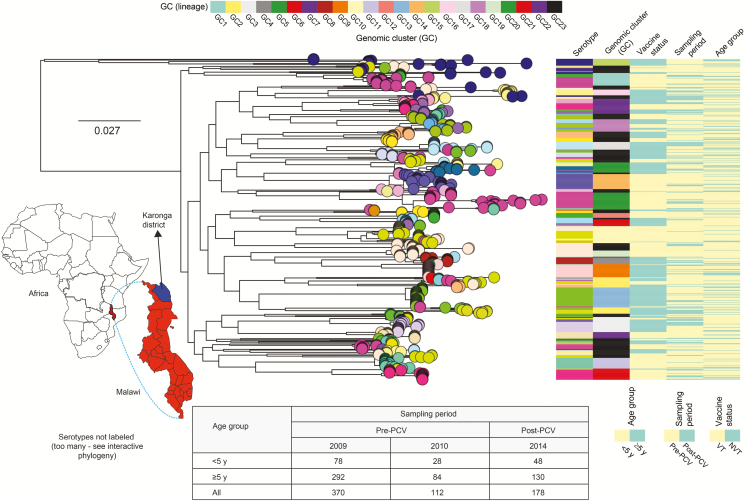
Figure 1.
Sampling location, genetic similarity, and distribution of carried pneumococcal isolates. The map of Africa shows the location of Malawi and Karonga district from which the isolates were sampled. The number of isolates (n = 660) used in the genomic analysis are shown in the table below the phylogenetic tree. The core genome maximum likelihood phylogenetic tree of the 660 carriage isolates rooted at the branch of “classical” nontypeables. The tips (circles) of the tree are colored by serotype, and colored panels to the right correspond to serotype, genomic clusters or lineage, vaccine status, sampling period, and age group. The tree with metadata and corresponding international definitions of the pneumococcal lineages is available interactively online at https://microreact.org/project/xH7-VcoWj/8a339d57. Abbreviations: GC, genomic cluster; NVT, nonvaccine serotype; PCV, pneumococcal conjugate vaccine; VT, vaccine serotype.