Fig. 3.
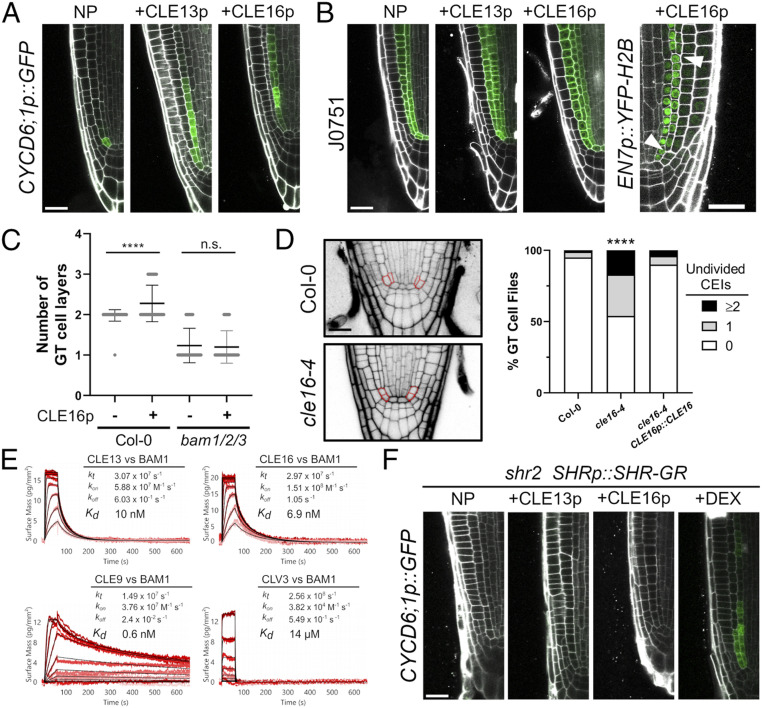
BAM-CLE signaling regulates SHR-dependent cell division. Expression of CYCLIND6;1 (A) and J0571 (B) in Col-0 roots with no peptide (NP), CLE13p, or CLE16p treatments. Expanded CYCLIND6;1p::GFP expression in Col-0 was observed when treated with CLE13p (n = 6/8) and CLE16p (n = 28/33) compared to no peptide controls (n = 0/23). (B) The EN7 marker becomes restricted to innermost cells following ectopic CLEp-induced asymmetric divisions (white arrowheads). (C) Col-0 roots show an increase in the number of ground tissue layers while bam1/2/3 are not affected by CLE16p treatment [n = 50, Col-0(−); n = 54, Col-0(+); n = 64, bam1/2/3(−); and n = 65, bam1/2/3(+)]. Distributions were compared using a Mann–Whitney nonparametric t test (****P ≤ 0.0001). (D) CEI division defects in the cle16-4 mutant are fully restored with CLE16p::CLE16 (n = 127, Col-0; n = 118, cle16-4; and n = 152, cle16-4 CLE16p::CLE16). Distributions were compared using a Kruskal–Wallace nonparametric test (****P ≤ 0.0001). (E) Quantitative binding kinetics of CLE peptides versus the BAM1 ectodomain by GCI. Shown are sensorgrams with raw data in red and their respective fits in black. Binding kinetics were analyzed by a one-to-one binding model with mass transport. Table summaries of kinetic parameters are shown: kt, mass transport coefficient; kon, association rate constant; koff, dissociation rate constant; and Kd, dissociation constant. (F) Expression of CYCLIND6;1 in shr2 mutants with NP, peptide treatments, or DEX control treatment. (Scale bar, 25 µm in A, C, and E.)