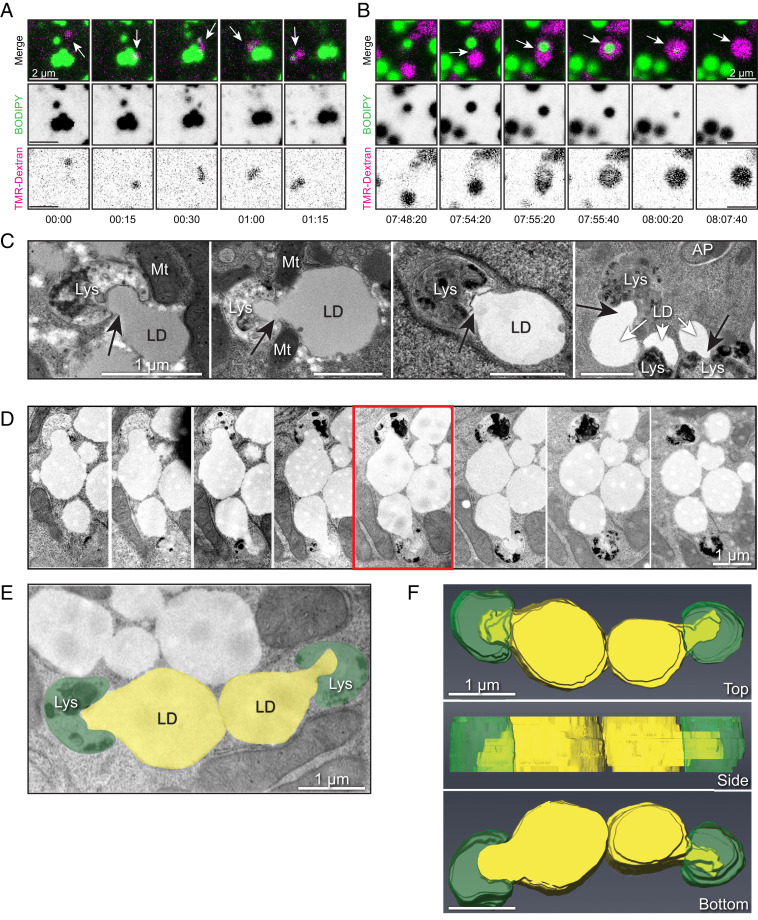
Fig. 3.
Direct interactions between LDs and lysosomes drive piecemeal lipid transfer. (A) Live-cell confocal imaging of primary rat hepatocyte incubated with fluorescent dextran (magenta) to label lysosomes and BODIPY-FL-C12 (green) to label LDs. Panels show time series stills of an example of a lysosome contacting an individual LD and pulling away a portion of fluorescent lipid signal, suggestive of direct lipid transfer between compartments. (B) Live-cell confocal imaging of a primary rat hepatocyte labeled as in A, showing an example of a lysosome directly enveloping and quickly degrading the internalized LD within less than 1 min. See corresponding Movie S4. (C) Additional electron micrographs of primary rat hepatocytes subjected to culture in media containing 150 μM oleic acid overnight (to stimulate LD formation) and serum-starved in HBSS (to promote LD catabolism) for 4 h before fixation and processing for TEM. Note the appearance of profiles of piecemeal degradation of the lipid droplet that appear to be directly transferred into adjacent degradative compartments. (All scale bars, 1 μm.) (D) Serial section electron microscopy through an instance of putative lipid transfer structures in primary rat hepatocytes loaded overnight with 150 μM oleate and subjected to 4-h HBSS starvation. Sections are ∼100 nm apart in successive panels. (E and F) The 3D representation of the serial sections used in D, generated using Amira Software (Thermo Scientific). Lipid droplets are pseudocolored yellow, while degradative structures are pseudocolored green (E). Note the apparent protrusion of lipid content into the adjacent degradative structures from a 3D reconstruction of a series of sections taken through the midplane of two LDs (F).