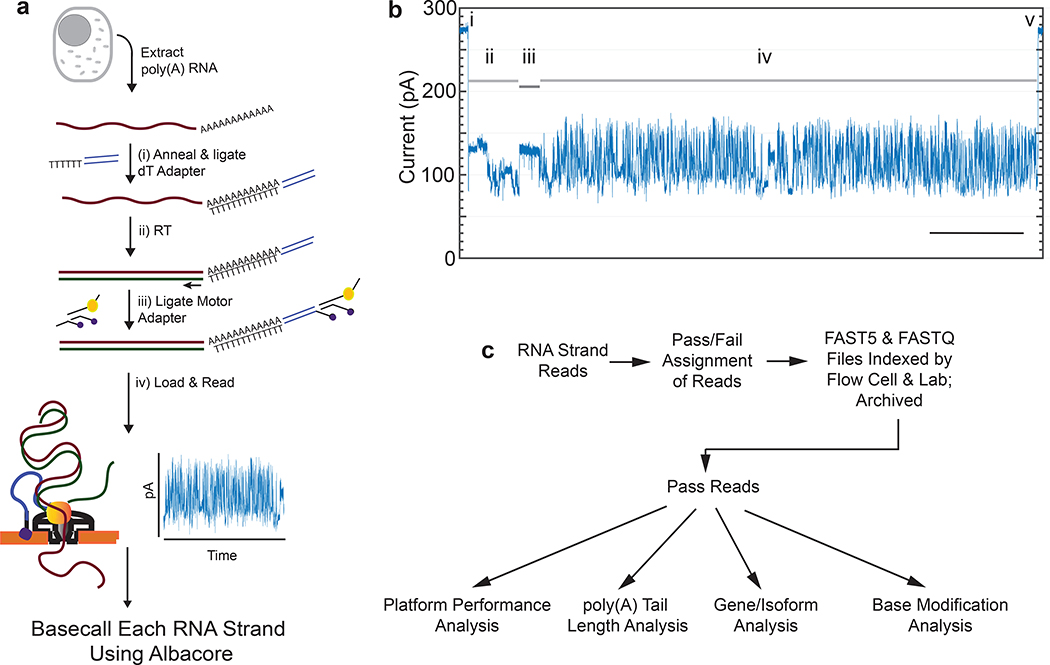
Figure 1.
Nanopore native poly(A) RNA sequencing pipeline. (a) RNA is isolated from cells followed by poly(A) selection using poly(dT) beads. Poly(A) RNA is then prepared for nanopore sequencing using the following steps: (i) A duplex adapter bearing a poly(dT) overhang is annealed to the RNA poly(A) tail, followed by ligation of the strand abutting the poly(A) tail; ii) the poly(dT) complement is extended by reverse transcription. This step improves throughput, but it is not necessary, and the cDNA strands are not read; iii) a proprietary ONT adapter bearing a motor enzyme is ligated to the first adapter; and (iv) the product is loaded onto the ONT flow cell for reading by ionic current impedance. The ionic current trace for each poly(A) RNA strand is base called using a proprietary ONT algorithm (Albacore). (b) A representative ionic current trace for a 2.3 kb TP3 transcript. Ionic current components: (i) Strand capture; ii) ONT adapter translocation; iii) poly(A) RNA tail translocation; iv) mRNA translocation; and (v) exit of the strand into the trans compartment. Bar is 5 seconds. (c) Processing of the RNA strand reads in silico, followed by data analysis.