Figure 5.
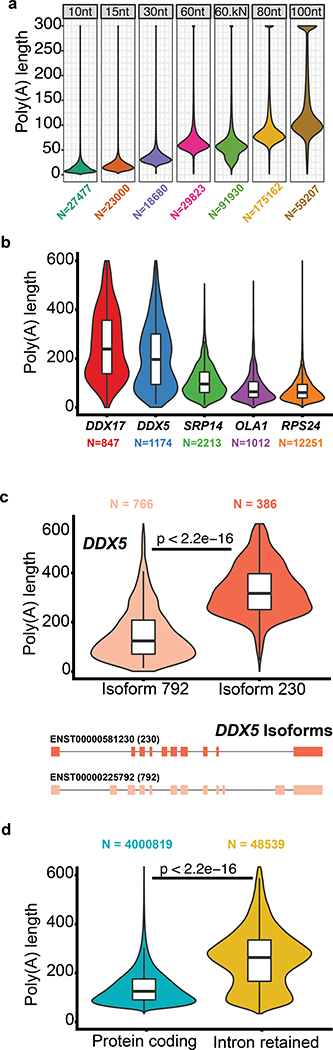
Testing and implementation of the poly(A) tail length estimator nanopolish-polya. (a) Estimate of poly(A) lengths for a synthetic enolase control transcript bearing 3′ poly(A) tails of 10, 15, 30, 60, 80 or 100 nucleotides. ‘60kN’ contained all possible combinations of a 10nt random sequence inserted between the enolase sequence and the 3′ poly(A) 60mer. (b) Violin/box plots showing poly(A) tail length distributions for genes with the longest (DDX5, DDX17), median (SRP14), and shortest (RPS24, OLA1) values from a ranked list of 1043 genes. (c) Distribution of poly(A) tail lengths and gene models for two isoforms of DDX5. (d) Distribution of poly(A) tail lengths for representative intron-retaining and intron-free transcripts identified using the GENCODE-Sensitive isoform set. Kruskal-Wallis p-value are denoted.