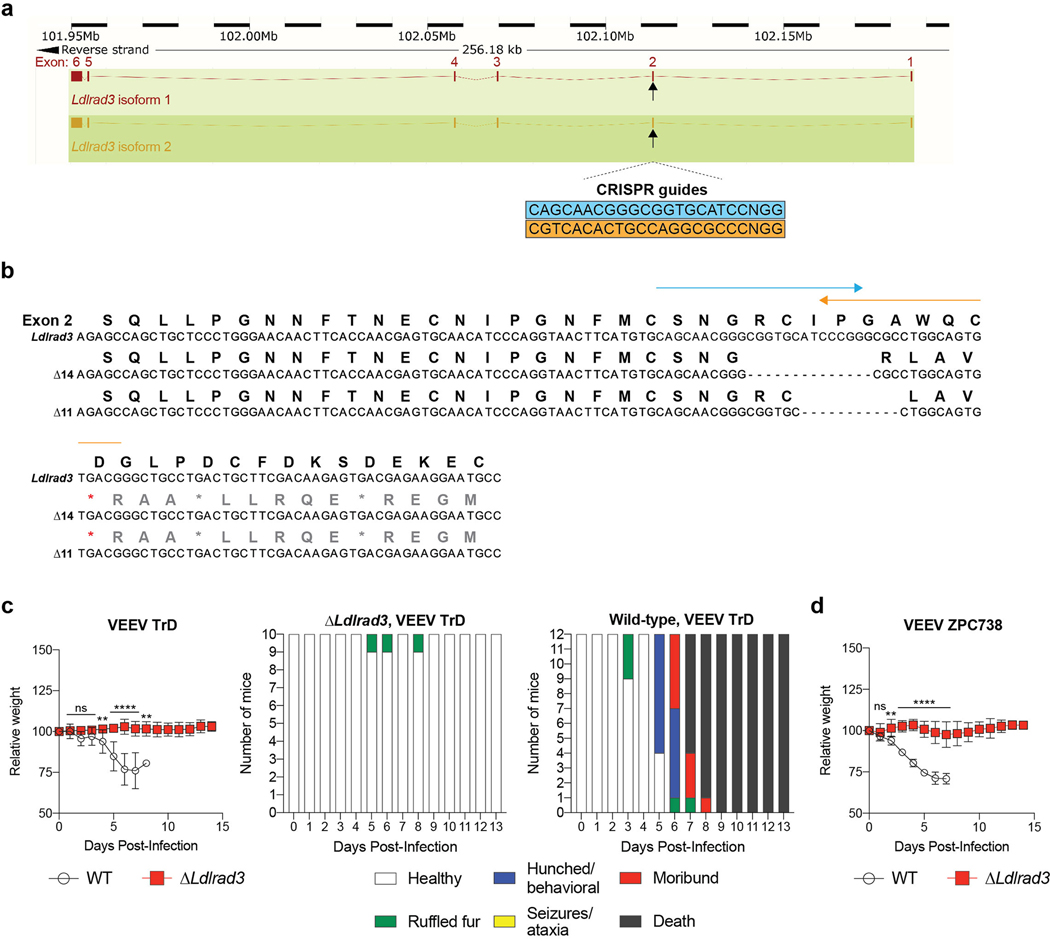
Extended Data Figure 9. Generation and clinical assessment of C57BL/6 mice with deletions in Ldlrad3 by CRISPR-Cas9 gene targeting.
a. Scheme of Ldlrad3 gene locus with two sgRNA targeting guides for a site in exon 2 of both isoforms. The full-length and truncated Δ32 N-terminus residue Ldlrad3 isoforms are colored red (top) and orange (bottom), respectively. b. Sequencing and alignment of Ldlrad3 sgRNA targeting region in exon 2 (11- and 14-nucleotide frameshift deletions) in gene-edited Ldlrad3 mice. The amino acid residues and the two sgRNA guides used for gene-editing (blue and orange arrows) are indicated above. c-d. Seven-week-old male and female mice with deletions in Ldlrad3 (Δ11 or Δ14 nucleotides; homozygous or compound heterozygous) or wild-type C57BL/6 mice were inoculated subcutaneously with 103 PFU of VEEV TrD (c, left panel) or 102 FFU of VEEV ZPC738 (d). Mice were monitored for weight change. Data are from two experiments (VEEV TrD: WT, n = 12; ΔLdlrad3, n = 10; VEEV ZPC738: WT, n = 9; ΔLdlrad3, n = 8; two-way ANOVA with Dunnett’s post-test: ** P < 0.01, **** P < 0.0001; n.s., not significant). c: 1 dpi, P > 0.999; 2 dpi, P = 0.2136; 3 dpi, P = 0.5489; 4 dpi, P = 0.0065; 8 dpi, P = 0.0014. d: 1 dpi, P = 0.8383; 2 dpi, P = 0.001. Clinical disease (right panels) was assessed over time (healthy, ruffled fur, hunched/behavioral, seizures/ataxia, moribund, or death) in mice inoculated with VEEV TrD (c, right panels).